The Genetic Research Methods and Its Role in Aquaculture on Indonesia
1Department of Aquaculture, Faculty of Fisheries and Marine Science, Pattimura University, Ambon, Indonesia
2Maritime and Marine Science Center for Excellence Pattimura University, Ambon, Indonesia
- *Corresponding Author:
- Laimeheriwa Bruri Melky
Department of Aquaculture
Faculty of Fisheries and Marine Science
Pattimura University, Ambon, Indonesia.
Tel: +62 911 322628
E-mail: bruripenabur@gmail.com
Received date: January 09, 2018; Accepted date: April 27, 2018; Published date: May 02, 2018
Citation: Laimeheriwa BM (2018) The Genetic Research Methods and Its Role in Aquaculture on Indonesia. Insights Aquac Cult Biotechnol Vol.2 No.1:3.
Abstract
Successful development of aquaculture was largely determined by the seed supply of both the quality and quantity of the seed. Seed quality was influenced by the quality of the genetic factors and environmental factors i.e. water quality, food, and disease. Improved parent quality through the improvement of both qualitative and quantitative genetic traits that could be done through the selection of traits or characteristics of prospective mothers, as well as avoiding the occurrence of inbreeding that causes heritabilities decline. The purpose of writing to review the results of genetic studies that have been applied in the field of aquaculture in Indonesia. In this article, the study included the following: basic genetic concepts and theories, genetic research methods, and their roles and functions in the field of aquaculture.
Keywords
Genetic research; Aquaculture genetic; Performance; Gynogenesis; Androgenesis; Heritability; Breeding value; DNA marker
Introduction
Aquaculture was any form of activity to produce biota or aquatic organisms in a controlled environment and in nature in order to obtain production or output and benefits or outcomes. Activities carried out in the sea, brackish waters, fresh water, including the general waters of the lake, reservoirs and rivers. In principle, aquaculture activities were to control growth, death, and reproduction.
Observing the status and potential of our nation's aquaculture, although overall national fishery production was still dominated by capture fisheries, the contribution of aquaculture has a much higher production growth compared to capture fisheries. Data from the Ministry of Marine Affairs and Fisheries 2018 shows that the contribution of aquaculture to national production increased from 70.07 percent in 2015 to 74.03 percent in 2017. In contrast, the contribution of capture fisheries decreased from 29.93 percent in 2015 to about 25.97 percent in 2017 [1]. This gives the impression that aquaculture in the future will play an increasingly important role, but it certainly must be accompanied by some notes of improvement of the weakness that has been found.
Domestic and world demand for fishery products continues to increase as population increases and human awareness of the benefits of healthful and educated fish increases. The capability of producing fishery products from capture fisheries activities at the global level was maximum of 100 million tons per year. The FAO data in 2018, the total volume of capture fisheries production was estimated at 94 million tons in 2025, up from about 93 million tons on average between 2013 and 2015 [2]. The main reason for the stagnation in this capture fishery production was that most fish stocks waters have been fully exploited, and now its catch quantity tends to decrease.
The potential of aquaculture production owned by Indonesia was about 57.7 million tons per year, the largest in the world. While in 2017 the total production of national aquaculture was only 17.22 million tons or 29.8 percent [2]. Thus, if aquaculture was more excavated and empowered through change and improvement, it was not impossible that the future will become a mainstay of the economy and increase the effect of a very large domino again. In the sense of providing opportunities for economic growth, job creation, and reducing poverty.
More than 90 percent of the world's aquaculture production comes from Asia, so it was only natural that Asian countries were the most likely to attract foreign exchange from aquaculture business. Indonesia has the opportunity to become a world leader in aquaculture as well as a mainstay of the economy. Moreover, the government's attention to this sector was increasingly evident with the big vision of making Indonesia sovereign in the maritime field by making Indonesia as the world maritime pivot.
Successful development of aquaculture is highly determined, among others: by the supply of seeds that include the quality and quantity factors of seed. Seed quality was influenced by the quality of the mother as genetic factors and environmental factors, i.e., water quality, food, and disease. The quality of the parent as a genetic quality factor has an influence on the quality of the seeds in terms of increasing the number of seeds, rapid growth, and resistance to environmental changes. Improving the quality of the mother through the improvement of both qualitative and quantitative genetic traits that chould be done through selection of traits or characters of prospective mothers. In addition, the management of the parent to avoid the occurrence of inbreeding that causes decreased heritability the parent.
This manuscript aimed to review the results of genetic studies that have been applied in the field of aquaculture. In this article, the study includes basic genetic concepts and theories, genetic research methods, and the role and function of genetic analysis in aquaculture.
The Basic Genetic Concepts and Theories
Although people usually establish genetics beginning with the rediscovery of the manuscript of an article written by Gregor Mendel in 1900, genetics as "the science of inheritance" or heredity has been known since prehistoric times, such as the domestication and development of various breeds of cattle and cultivars [3]. People have also known the effects of crossing and close marriage and made a number of procedures and regulations about it since before genetics stood as an independent science [4]. Genealogy of the disease in the family, for example, has been reviewed before. However, this practical knowledge does not explain the cause of the symptoms.
The popular theory of inheritance adhered to in that period was the theory of inheritance: one inherits a flat mixture of attributes brought by the elderly, especially of the males for carrying sperm. Mendel's results show that this theory was not applicable because the properties were carried in combinations carried by typical alleles, rather than mixed blends. Another related opinion was Lamarck's theory: the traits that the elder inherited in his life be passed on to his son. This theory was also broken with Mendel's explanation that the nature brought by the gene was not influenced by the experience of the individual who inherits that trait. Charles Darwin also gave an explanation of the pangenesis hypothesis and later modified by Charlesworth and Charlesworth [5]. In this view, the body's cells produce particles called gemmules that will be collected in the reproductive organs before fertilization occurs. Thus, every cell in the body has a contribution to the properties that its offspring [6].
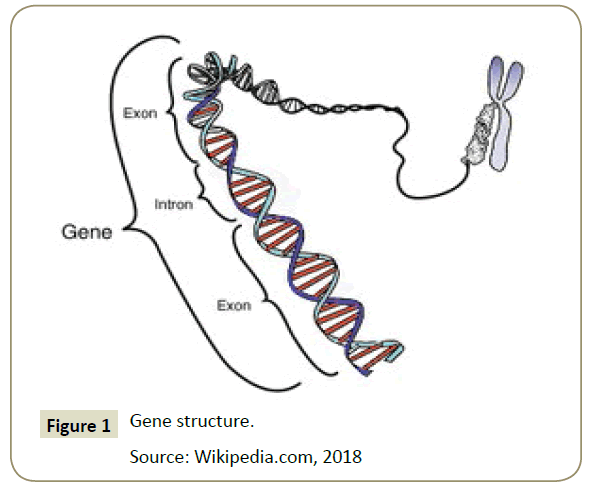
Gene, from the Dutch: genes, were the inheritance units of nature for living organisms [7]. The physical form was a sequence of DNA encoding a protein, polypeptide, or an RNA that has a function for the organism that has it. Modern gene boundaries were a particular location in the genome that was related to inheritance properties and chould be associated with functions as regulators or controllers, transcription goals or other functional roles [5,8]. The use of "genes" in everyday conversations, eg "smart genes" or "hair genes", were often meant for alleles: the choice of variations available by a gene [9] (Figure 1). Although the allele's expression chould be similar, people more often use the term allele for expression of genes that were phenotypically different [10]. The genes were inherited from one individual to their offspring through a process of reproduction, together with the DNA that carries it [11]. Thus, information that maintains the integrity of the form and function of an organism's life chould be maintained.
At that time DNA has been discovered and was known to reside only on chromosomes, but people have not yet realized that DNA was linked to genes [12,13]. Through Oswald Avery's study of Pneumococcus bacteria, and Alfred Hershey and Martha Chase with T2 bacteriophage virus, did people know that DNA was a genetic material. In the 1940s, George Beadle and Edward Tatum experimented with Neurospora crassa. From these experiments, Beadle and Tatum chould draw hypotheses that genes encode enzymes and they conclude that one gene synthesizes one enzyme or one gene-one enzyme theory [14]. A few decades later, it was discovered that genes encode proteins that not only function as enzymes and some proteins were composed of two or more polypeptides [15,16]. With these findings, Beadle and Tatum's opinion, one gene-one enzyme theory, has been modified into the theory of one gene-one polypeptide or one gene-one polypeptide theory.
In eukaryotic cells, the gene consists of the transcriptional initiation regulatory domain, comprising the following: GCCACACCC series, ATGCAAAT, GC boxes, CCAAT boxes and TATA boxes, introns and exons that were encoded protein areas that chould be transcribed overlapping or nonoverlapping [17]. For example, in a code with three series of nucleotides, as triplet codons, i.e., AUU GCU CAG, it chould be read nonoverlapping as AUU GCU CAG or read overlapping as UGC UUG UGC GCU CUC CAG. Although in about 1961, it has been found that amino acids coded by codons by nonoverlapping have found different proteins transcribed with shifting overlapping codons, as well as the final regulatory domain of the transcription [18].
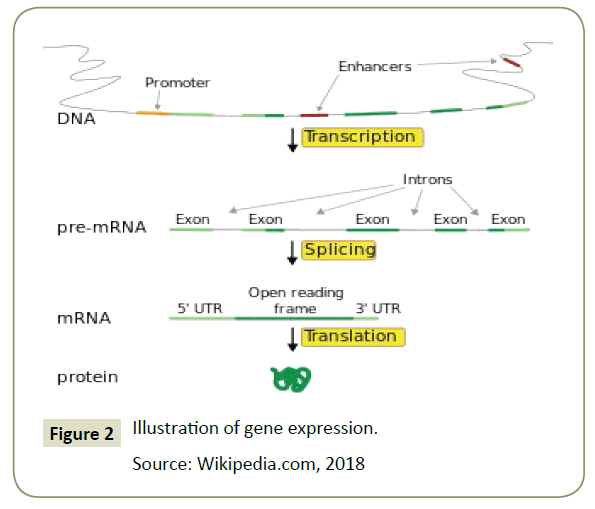
Gene expression was the process by which the information codes present in the gene were converted into proteins that operate only within the cell [10]. Gene expression consists of two stages: the process of making RNA copies or transcription and polypeptide synthesis processes specifically in the ribosome or translational. The process of transcribing DNA into mRNA and translating mRNA into a polypeptide was called the central dogma. The central dogma applies to prokaryotes and eukaryotes [19]. However, in eukaryotes, there was an additional stage that occurs between transcription and translation called the premRNA stage. The pre-mRNA stage was to select the mRNA to be sent out of the nucleus to translate in the ribosome [20]. The exon was the mRNA to be sent out of the nucleus to be translated, while the introns were the mRNAs that will remain in the nucleus because it was likely that the mRNA will form a protein that was not functional if it was translated. Intron will then break down again to form a new mRNA chain. It was also known that some of the so-called mutations chould occur in the process of expression of this gene [21] (Figure 2).
Genotype, literally "gene type", was the term used to express the genetic state of an individual or a group of individual populations [22]. Genotypes may refer to the genetic state of a locus as well as the entire genetic material carried by the chromosome or genome [23]. Genotypes chould be homozygous or heterozygous. Once people chould transfer gene, there was also the use of the term hemizygote.
In Mendel's genetics, as classical genetics, genotypes were often denoted by pairs of letters; such as AA, Aa, or B1B1. The same pair of pairs indicates that the individual symbolized was homozygous, i.e., AA and B1B1, while the different letter pairs represent the heterozygous individual. A pair of letters indicates that this symbolized individual was diploid or 2n [24]. As a consequence, for example, the individual tetraploid or 4n homozygote was denoted by AAAA [7].
The phenotype was a characteristic, both structural, biochemical, physiological, and behavioral, that chould be observed from an organism regulated by genotype and its environment and interaction [25]. The phenotypic definition includes the various levels of the gene expression of an organism. At the organism level, a phenotype was something that chould be seen or observed, something of nature or character. In this stage, phenotypes such as eye color, weight or resistance to a particular disease. At the biochemical level, the phenotype may be the content of certain chemical substances in the body [26]. For example, blood sugar or protein content in rice. At the molecular level, the phenotype may be the amount of RNA produced or the detection of DNA or RNA bands in electrophoresis.
Phenotypes were determined in part by individual genotypes, in part by the environment in which the individual lives, time, and, in some traits, the interaction between genotype and environment [27]. Phenotypic observations can be simple - usually flower color - or very complicated to require special tools and methods [28]. However, due to the genetic expression of a gradual genotype from the molecular level to the individual level, there was often a link between a number of phenotypes at different levels.So far the discussion of a phenotype was assumed to describe its genotype. Phenotypes of such traits were easy to distinguish, for example, the color of the bean husk was alternatively white or gray, the plant was high or low, so it was easy to distinguish. Such properties were known as discontinuous traits [29,30]. In such traits, there was a simple relationship between genotype and phenotype. In many cases a genotype produces only one kind of phenotype, otherwise, a phenotype was the result of the activity of a genotype. Nevertheless, we have come to understand that the relationship between genotype and phenotype was influenced by phenomena, such as expressivity, penetration, and pleiotropy [31]. In other words, a genotype chould produce different phenotypes because the genotype interacts with its environment during the growth and development process. The properties such as fish weight, fish length, and protein content show a wide range of phenotypes.
The properties that indicate such a range of phenotypic diversity were often called continuous traits [32]. On discontinuous properties have a discrete distribution, whereas continuous properties have a continuous distribution, so it must be expressed quantitatively [33]. Such properties were often called quantitative properties. Continuous properties have a continuous range of phenotypes as well. To examine the inheritance of the continuous or quantitative traits it was necessary to see why a trait has various phenotypes [34]. Quantitative properties chould occur in various ways. Generally, the phenotype range occurs because different genotypes exist in an individual group or population. This was common when a character was controlled by many loci. For example, if a locus with two alleles per locus controls a trait, then there were three possible genotypes namely AA, Aa, aa. For two loci with two alleles per locus, there will be 9 genotypes, eg AABB, AABb, Aabb, AaBB, AaBb, Aabb, aaBB, aaBb and aabb. Similarly, the increase in the number of loci as much as n, then the genotype formed as much as 3n. if there were more than two alleles for each locus it will produce much more genotype.
Thus the more the locus that controls a trait, the more likely genotypes may be formed. The properties controlled by many such loci were called polygenic or quantitative properties. polygenic or quantitative properties [35,36]. The basic difference between qualitative and quantitative properties involves the number of genes that contribute to the phenotype variability and the degree to which the phenotype chould be modified by environmental factors [36]. Quantitative properties chould be governed by many genes (perhaps 100 to 100 or more), each contributing to the phenotype so little that its individual influences chouldnot be detected by Mendel's methods. Such genes were called polygenic.
Below was presented a summary of some major differences between quantitative and qualitative genetics (Table 1). In the inheritance of quantitative properties, the concept of polygenic was used to explain the formation of quantitative traits [37]. Ronald Fisher chould explain that the quantitative traits was formed of many genes with little influence, each of which segregates according to Mendel's theory [38-41]. Because of its small effect, the phenotype regulated by these genes chould be affected by the environment. Nevertheless, Fisher's explanation still places the "genes" that govern quantitative properties as abstract because they were only concepts [37,41]. The proof of the existence of genes that regulate quantitative properties begins to open up after the availability of many genetic markers to enable people to create genetic maps that chould reach most of the chromosomes. Genetic markers were used to indicate the allelic situation in a particular chromosome part [42]. Allele variations on a marker become genotypes for chromosomes or clusters.
| Num. | Quantitative Traits | Qualitative traits |
|---|---|---|
| 1. | Characteristics of the degree | The characteristics of the type. |
| 2. | Continuous variation; phenotype measurement is a spectrum | Discontinuous variations; clear phenotype classes |
| 3. | Polygenic control; the influence of single genes is too small to be detectable | The single gene gives a distinctly differentiated influence |
| 4. | Questioning a population of organisms comprising all kinds of marriages that chould occur | Questioning the marriages of individuals and their descendants |
| 5. | Statistical analysis provides estimates of population parameters such as averages and standard deviations | Analyzed by making calculations and ratios |
Table 1. Main differences in inheritance of quantitative and qualitative traits.
Heritability was the proportion of genetic variation to the total amount of genetic variation coupled with the variety of environment, in other words, heritability was the proportion of the genetic variation to the phenotype variety for a particular character [43,44]. There were two heritability values known in plant breeding that were heritability in the broad sense and heritability in the narrow sense [45].
The value of heritability in the broad sense takes into account the total genetic variation in relation to phenotype diversity. In this case, the genotype was considered a unit in relation to the environment [43,44]. While narrow heritability was the focus of attention, the diversity attributed to the role of additive genes was part of the total genetic diversity. Based on this explanation it chould be understood that the value of heritability in the narrow sense will never be greater than the value of heritability in a broad sense for a particular character.
Heritability parameters involve all types of gene action and therefore form a broad heritability estimate [46,47]. In the case of perfect dominance, if a gamete containing the dominant active allele A2 combines with a gamete containing a zero allele A1, the resulting phenotype may consist of two units [47]. When two A2 gametes combine, the phenotype results will consist of two units. Conversely, if genes that have no dominance or additive genes were involved, then the A2 gametes will add one unit to the phenotype of the resulting zygote, regardless of the contribution of the allele of the gamete to which it combines. Thus only the genetic additive component of the variance has predictable quality, which was in the formulation of breeding roles. Heritability in this narrower sense was the ratio of genetic additive variance to the phenotype variance [48,49].
Genetic Research Methods
Phenotypic performance analysis
Phenotypic performance analysis includes morphological and morphometric data. Measured morphometric data include weight, length, width, height, shell thickness, area and circumference [36,50]. The morphometric ratio or index analysis was performed to obtain the coefficient of interpersonal performance within a population by calculating the distance coefficient or Eucleudian value, the coefficient of similarity and coefficient of variation. The coefficient values were mapped in the dendrogram to see whether there were similarities and differences between individuals in the ring snail population [51].
Biochemical analysis method
The measured biochemical performance was the analysis of enzyme or allozyme, protein, amino acids using starch gel electrophoresis [52,53]. The analysis of the buffer migration rate and the staining procedure [54,55]. Fitness test for Hardy- Weinberg equilibrium and differences between populations using GENEPOP [56]. The interpretation of locus and allele identification follows the Allendorf and Utler method. The allele's naming was based on the relative distance of the migration of the allele to the most commonly encountered allele coded 100. The data was interpreted to obtain allelic frequency, the degree of polymorphism, heterozygosity and genetic distance.
Discriminant analysis was used to see the significant differences between groups determined by observed morphometric. The measured results were relativized using standard deviations to avoid the diversity of sizes and possibly different ages. Next will be grouped morphological characters that most characterize the difference. The influence of the interaction of fellow characters measured resulted in the depreciation of these characters. This component was also used to see the most powerful influence on the formation of a population. This analysis uses Minitab program and SPSS program based on Principal Component Analysis or PCA [57-59]. PCA analysis aims to interpret most of the information contained in a data matrix into graphs and to obtain maps of population distribution and similarity values within and outside the group. The results obtained were shown in dendrogram form.
RNA and DNA ratio analysis
Total DNA and RNA extracts follow the standard method developed [60,61]. Amplification and visualization of DNA and RNA using PCR and electrophoresis methods with primers tailored to the sample [62]. DNA sequencing and RNA from PCR using Sequenzer ABI377A Applied Biosystem. The DNA sequence and RNA were edited manually using Genetyx software, then aligned using Clustal software [63,64]. Characterization of DNA sequences and their analysis using the maximum parsimony and Neighbor Joint methods performed using MEGA software [65]. To find out the concentration of DNA and RNA measured by using Gene quant program. Results of DNA and RNA genomic concentrations of each sample were further calculated for determination of RNA-DNA ratio [66,67].
DNA marker methods
Some DNA markers that can be used to analyze genetic variation, the evolution of a plant, the linking of specific genes to specific characters, parent search, analysis of quantitative character loci and revision of plant classification were Restriction Fragment Length Polymorphism, Random Amplified Polymorphic DNA, Amplified Fragment Length Polymorphism and microsatellite or Simple Sequence Repeat [68,69].
Restriction fragment length polymorphism (RFLP)
Research to exploit DNA polymorphisms in plant genomes by utilizing the technology of molecular markers continues to increase. RFLP was a molecular technique based on polymorphism caused by the substitution of the base or nucleotide, insertion, dilation, or translocation that may occur in the past [70]. This technique utilizes specific restriction sites in the genome of an organism. Restriction enzyme or restriction endonuclease was an enzyme that cuts DNA strands on a sugar-phosphate framework without damaging a base, in the sequence it recognizes. The recognition sequence was often called the introduction site was a DNA sequence that places the restriction enzyme and cuts on the sequence. These enzymes can be ordered through companies engaged in biotechnology and molecular biology. The result of genomic cutting using certain restriction enzymes will result in differences in the length of the DNA fragment, which shows the distance from the enzyme restriction sites in an organism's genome. The fragmented genome could then be analyzed for the purpose of the study. Through further analysis could be known whether the target sequence has been changed due to the substitution of the nucleotide, insertion, deletion or translocation.
Random amplified polymorphic DNA (RAPD)
RAPD was widely used to analyze the diversity of genetic traits in a variety of studies with the consideration that, among others, it does not require a background knowledge of the genome to be analyzed, primers used universally - could be used for both prokaryotes and eukaryotes-, capable of producing relatively unlimited characters, the materials used were relatively cheaper, the preparation was easier, and yields faster results than other molecular analyzes [71,72]. The RAPD method was able to detect the nucleotide sequence by using only one primer. The primer will bind to the single strand of the one genome and to the DNA strand of its partner in the opposite direction [73]. As long as the primary attachment site remains at an amplified range in general no more than 5000 bp, it will obtain amplification DNA products. With the use of RAPD that was relatively easy, inexpensive and analyzed in DNA level, early selection could be performed on the properties of resistance to biotic and abiotic stress, quantitative characteristics and duplication of collection crops could be avoided [73]. Random Amplified Polymorphism DNA was one of the most widely used molecular techniques used in the study of the population's genetic structure and measurement of the genetic variation of a population of organisms. The number and distribution of genetic variations reflect the interaction between the processes involved in evolution such as genes, mutations and genetic drift.
Microsatellite (Simple sequence repeat, SSR)
Repeated recurrent DNA that has the highest variation in the plant genome was a recurrent sequence with a simple or short repetitive fragment [74]. This fragment was known by the name minisatellite and microsatellite. Minisatellite was a repetitive DNA usually between 10-60 bp, which at the beginning of its discovery was widely applied to the human genome [75]. While SSR has fewer recurring units ranging from 1 to 6 bp, contained in very large quantities and spread in the genome and was widely used in plants. Variations of these fragments are usually the result of changes in the number of copies of the original loop and were often categorized as a variable number of tandem repeats or VNTR [76,77]. Because very high levels of polymorphism could be detected with this fragment, VNTR was recognized as a potent tool for fingerprinting and cultivar identification. This fragment could also be used to study the diversity between and intra-population, ecological studies, calculating genetic distance and studying plant evolution. The short sequence of DNA microsatellites with the clamp DNA sequences was conserved, allowing the primary design to amplify specific sites using PCR. If these primers were used to amplify certain SSR loci, then each primer will produce polymorphism in the form of a difference in amplification length known as Simple Sequence Length Polymorphism or SSLP. Each length represents one allele of a locus. Long differences occur because of the difference in the number of repeating units at specific SSR loci [74,75]. The diversity of the number of replicates in microsatellite could be detected by electrophoresing the amplified DNA products in the standard gel sequence, which could separate the fragments that distinguish each nucleotide. Microsatellite DNA was present in large quantities and spreads in the genome. The general form of repetition of DNA microsatellite was a simple repetition of two bases. Microsatellite DNA with the ease and speed of using PCR technology, co-dominant and easily interpreted makes microsatellite the best marker in gene mapping [78].
Molecular data analysis
Data generated from the molecular analysis were often used as a basis for the classification of individuals in a population, for phylogenetic construction, parent search, identification of markers of a particular trait, taxonomic revision and others [79- 81]. The dendrogram constructed in the analysis was generally based on the degree of similarity between the individuals used. However, the confidence limits for groupings generated through the dendrogram, usually not acceptable for calculations using common statistical procedures [82]. Molecular data to be analyzed should be converted into binary data based on the presence or absence of the amplified band. If there are amplified bands scored 1, whereas if none was given a score of 0. Binary data obtained could be used to arrange the dendrogram or phylogenetic of the organism being analyzed. For parent search, genetic mapping, and identification of specific fragments of binary data were converted into genotype data, and with specific programs analyzed according to the destination [80].
The result of the molecular band pattern that has been converted into binary data form could be processed using NtSys software to obtain the matrix of genetic distance and dendrogram or phylogenetic of the sample [70,83]. To analyze the data according to research objectives, there was now very much software that could be used to process molecular data. For example, Genotype data composed of binary data could be used to analyze the relationship of the parent to the filial, the linkage map, and the correlation between loci with locus and locus with morphological properties. Tracking the elders using Cervus, cross-based depression in molecular markers using POPGENE, making a linkage map using MAPMAKER software with LOD 3 values, and the correlation between loci with locus and between the locus and morphological properties using statistical software such as Excel Stat, Minitab and SPSS [84,85].
The Role and Function of Genetic Research in the Field of Aquaculture
Observation of aquaculture biota performance
Variation of phenotype as an indicator of character performance One of the factors that determine the productivity of aquaculture was the genetic potential possessed by the cultivated population. Populations that have good genetic potential and were supported by appropriate cultivation environments will produce better productivity compared to individuals or populations with poor genetic potential [51]. Therefore, the selection of individuals or populations with good genetic potential needs to be done in order to improve the productivity of an aquaculture activity [86]. One of the important parameters to be considered was the genetic variability coefficient of the population. The genetic variability coefficient was the index value that indicates the magnitude of the genetic diversity of a population. In a population with high genetic diversity, the opportunity to find individuals who have advantages over certain traits.
The performance or expression of a character or the nature of an organism has a unique or specific or special order pattern. By seeing, knowing, recognizing and analyzing these unique or specific patterns of order, the breeding, selection, management, and conservation of those organisms could be done more easily, both quickly and effectively and efficiently [87,88]. Performance characteristics or phenotype characteristics could be qualitative and quantitative. Qualitative qualities or traits are categorized or described traits, whereas quantitative traits or characteristics were measured traits rather than descriptive [89,90]. These characteristics do not separate individuals over different phenotypic categories, such as black or albino. In fact, every character or quantitative trait shows a continuous distribution or distribution in a population. The difference between the individual phenotypes was a matter of degree or degree of difference rather than its kind.
Since the quantitative phenotype shows continuous characters or variations, the only way to learn it was to analyze the variants or variations that exist in one population and sort into the component parts [87-90]. The variant or variety of phenotypes analyzed for a quantitative feature was the sum or combination of genetic or genotype variants, variants or various environments as well as the interactions between genetic variants and the environment. A genetic variant or variety was a combination or a result of an additive genetic variant, a variant or a dominant genetic variant and an epistatic genetic variant. By analyzing the qualitative and/ or quantitative phenotypes, it was desirable to find patterns of order in a performance of undiscovered quantitative traits [91,92]. In one population, the quantitative phenotypes were described by the central tendency as well as the distribution or distribution around the tendency. For example, mean, variance, standard deviation, co-variant and range.
The inheritance of the quantitative character was very complicated, unlike the qualitative phenotype that could be controlled by one or a pair of single genes. Quantitative phenotypes were controlled by 20 or 50 or even more than 100 genes. Even the number of genes that control the quantitative phenotype was sometimes unknown and the mode of gene expression was unknown. Whereas the inheritance of quantitative phenotypes, such as egg production, body size, body weight, growth and graduation of life, was of great importance in fish breeding efforts, both consumption fish and bait fish [87]. This was inversely proportional to the breeding of ornamental fish that prioritizes the qualitative phenotype such as the color and shape of the fish body.
RNA and DNA ratio as an indicator of growth and nutritional conditions
The expression of RNA and DNA content in individual cells could serve as an indicator of the growth rate of the individual, this was because in the cytogenetic theory of the main RNA function as a protein-forming in cells that changes with cell division while the amount and size of DNA in an individual was always constant. Furthermore, the RNA/DNA ratio could be used as an indicator to describe the nutritional quality conditions consumed by aquaculture biota [66,67,93]. Thus, RNA/DNA ratio analysis could be used to assess and evaluate the quality of aquaculture biota.
DNA marker methods to evaluation and diagnosis
RAPD was one of the molecular markers used to evaluate the genetic relationship of organism populations. Information on genetic diversity and individual relationships between species through the fingerprint maps of the Eucheuma spp. type seaweed from the wild and cultivated areas of Sukabumi, West Java have been done by RAPD PCR method [74].
The diagnosis of aquaculture biota could be done in several ways, including isolation of the disease-causing agent and its morphological analysis, antibody detection resulting from infection with enzyme-linked immunosorbent assay or ELISA techniques and gene detection of the disease-carrier agent with PCR [94-96]. One of the most widely applied and developed techniques today was PCR capable of amplifying DNA fragments in vitro. In its development, has been developed Real-time PCR technique that was able to evaluate and do the quantification directly [97,98]. This technique was done by integrating PCR techniques with computers and software. The result of DNA amplification with PCR conventional, observation of DNA existence done at end of the reaction by using gel agarose after electrophoresis process. While the analysis using Real-Time PCR allows for observation at the time of reaction, the presence of amplified DNA could be observed on graphs that appear as a result of fluorescence accumulation of the probe. In Real-Time PCR observations the results no longer require the electrophoresis stage, so it was no longer necessary for agarose gel and the use of Ethidium Bromide which was a carcinogenic compound. White Spot Syndrome detection method: the virus in crabs as a WSSV carrier that infects tiger shrimp using conventional PCR and Real- Time PCR has been done by Pranawaty et al. [99].
Estimating heritability as an indicator of productivity
Heritability was the proportion of population phenotypic variation caused by genetic factors [43,44]. The concept of heritability was intended to assess a quantitative character in relation to the relative contribution of genetic and environmental factors to a particular trait. For example, how far the effects of genes affect habits, eg drinking alcohol, this will be useful in making social policy but this needs to be interpreted with great caution because of the high possibility of abuse misinterpretation [46,48,100]. To assess a heritability, the first step to take was to quantify the phenotypic variation of a property, then proceed with the variance into various sources of the cause.
The superior genetic properties utilized in a cultivation business need to be controlled and maintained and maintained its genetic quality. Efforts to control and maintain superior genetic quality could be done through breeding activities [101,102]. One of the things to note was knowledge of the value of heritability. Heritability shows how much a genetic factor a population affects its offspring compared to environmental factors. The greater the value of heritability in the superior traits of a population, the greater it was to obtain offspring with superior properties such as their parents.
It should be emphasized that the heritability of a trait applies only to a particular population living in a specific environment. A genetically distinct population living in an identical environment was likely to have different heritabilities for the same trait [103]. Similarly, the same population was likely to exhibit different heritabilities for the same trait when measured in different environments, since a given genotype does not always respond to different environments in the same way. No single even genotype has superior adaptability in all environments. This was why natural selection tends tCelcius degree cause genetically distinct populations within a species, a population adapting typically to local conditions and not generally adapting to all environments in which it was found.
The question that always arises in quantitative traits was the question of how far a trait was genetically controlled and how far the portion of the control was by the environment. Thus the repetitive question was how the balance of natural versus nurture or genes versus environment influences. This was the focus of the discussion of quantitative genetics. How much variation a character or phenotype observed was caused by genetic variation and environmental variation. To measure a phenotype variation and sort it into genetic variations and environmental variations, statistical methods are used. Example of the importance of Heritability: Selection of papaya endurance character against anthracnose could be done by knowing the character related to resistance to anthracnose, looking for a genetic marker with correlation test between character, heritability prediction and gene action that plays role in controlling anthracnose resistance in papaya [104-106].
The heritability estimation was useful to know how big a character could be inherited. Heritability was the ratio between the magnitude of the genotype and the total number of phenotypes of a character [107,108]. The broad heritability value for the character of the total solid was soluble; the hardness of the fruit and the anthracnose resistance was high. So these three characters have great opportunities could be genetically derived. Widely heritability estimates or h2 for endurance characters, total soluble solids and fruit hardness were high and the narrow sense h2 was moderate for the character of resistance and total dissolved solids but was low for fruit hardness.
Apart from the benefits, the heritability parameters have various limitations that were usually ignored, resulting in misuse. Hence the concept of heritability was often referred to as a parameter most often misunderstood and misused in the genetic field. Below were some of the qualifications and limitations of the heritability concept to consider: (a) heritability does not imply that a trait was genetically controlled [109,110]. What was measurable from heritability parameters was the proportion of phenotypic variance among individuals in a population because of their genotype difference. This was often misunderstood and regarded as a genetically controlled trait. A gene often affects the development of a trait, so it could be said that the character was genetic. However, the difference in the phenotype between individuals in a population measured in heritability parameters was by no means genetic; (b) heritability was measured by the value of variance that could only be calculated for an individual group, so heritability was a characteristic of a population rather than an individual character; (c) heritability was not fixed for a character. There was no particular value that was common to a character. The heritability value of a character depends very much on the genotype and the environment in which the genotype grows; (d) even if there were two populations where each population has a high heritability value for very different characters, it could not be said that the two populations were genetically different; and (e) a character possessed by individuals in a family does not always have high heritability value. Such characters were called familial trait. Family characters could be caused by the same genes or by the same environment, so familiarity was not synonymous with heritability.
Selection and breeding program
Selection program
Selection was a program by which individuals or families were selected, in an attempt, to change the average population in the next generation or, in other words, the selection was to choose individuals to be potential elders of the next generation. Selection was usually based on a minimum level of the performance index, [22,111]. Fish or individuals that exceed the minimum performance index will be selected as the parent stock while individuals under the minimal index or performance will be discarded or ignored. This means that the ratio or indexes measured or calculated were the performance values of individuals in a population.
In a program of character selection, quantitative traits assessed or analyzed were what genetic variants were responsible for producing such characteristics or quantitative traits that must be transmitted from parent to child in a predictable and reliable way. That was, genetic variant predictions were produced in the form of ratios or indices among the emerging quantitative character traits. For example, to predict inheritance properties such as: (a) the amount of egg production or fecundity of a fish, then one of the quantitative traits or characteristics measured was the performance or index of gonad size and egg size [112- 114]. Therefore, what genetic traits that produce or cause or control the development of gonad size and fish eggs need to be measured or analyzed; (b) growth rate, then one of the quantitative characters seen or measured is the performance or index of the size of the stomach, the length and diameter of the intestine, the size of the mouth of the mouth, etc., because produce these traits there were various genes control or manage it; (c) life graduation or sensitivity to environmental conditions or homeostatic abilities, then one of the quantitative properties measured was the performance of the heart and other respiratory devices, because there was a genetic variation that produces or controls it; and (d) the ability or efficiency of food conversion, the predictable quantitative trait was the digestive apparatus or channels involved in this digestion process, and all these were controlled by a number of even the many different genes involved.
This means the variety or variant of the size of the stomach, gonads, heart, intestine and others was an expression of the genetic variation that controls and produces it.Of the three variants or genetic variants of the additive variant, the dominant variant and the epistatic variant, only variants or additive variants could be transmitted in predictable and reliable ways, since the variety of additives was the function of allele, while the dominant variety was a genotype function that was the interaction between an inter alella at each locus [115-118]. So that additive diversity was sometimes regarded as a determinant factor in the breeding values of an individual. This means that the value of performance or index-index used was considered as a variety of additives. Whether the minimum or maximum spreads that will be used as a benchmark depend on the objectives to be achieved, such as whether for ornamental fish or consumption fish, whereas the mean or even mode was the frequency of the most frequent or frequent traits that could be used to predict or predict the genetic variants of the individual [119,120].
The individual deviations seen from the performance of cardiac indexes, stomach, meat, bone, body weight, body height, etc. could be used as additive variants which were further compared or converted to the ideal size or character could be used as parameter for selection program which then sought genetic distance and cluster [121,122]. In addition, the performance of the indexes for selection should be attempted on quantitative traits that could be measured at the time of the fish or the individual's life and not when it was switched off. For example, measurement of fin indexes, eye diameter, head length, body size, body weight and others.
Breeding program
Breeding was the application of biology, especially genetics, in the field of livestock to improve production or quality [123,124]. This science was relatively new and was born as the implication of the development of human understanding of the principles of inheritance of genetic traits. In general, this science seeks to explain and apply genetic principles in breeding activities. In practice, livestock breeding applies genetics, statistics, and biometrics as well as livestock reproduction, with the aim of improving the genetic quality of livestock, so as to increase production or provide added value in its implementation [125,126]. From the science of genetics, related to the aspect of decreasing the nature of the elder to his offspring. Included in this are Mendel's legal concepts. Statistics and biometrics play a role in measuring the diversity of traits and their distribution, the relationship between two or more traits, as well as analyzes for estimating genetic parameters. Reproduction was associated with aspects of fertility, fortune, breeding distance and birth [117,127,128].
Selection was a program by which individuals or groups were selected, in an attempt to change the average population in the next generation or, in other words, the selection was to choose individuals to serve as future elders of the next generation. Selection was usually based on a minimum level of performance or index. Fish or individuals that exceed the minimum performance or index will be selected as the parent stock while the individual under the index or minimum performance will be discarded or ignored [118,120,129]. This means that the ratio or index of the index measured or calculated was the value of the performance of individuals in a population. Selection was done in the hope that the selected individual or prospective elder will inherit a population with mean and range or range similar to the value contained in the selected individual compared with the mean and the distribution range originally. This means that the mean could be regarded as the largest frequency or range that was used as a benchmark inheritance of a genetic character.
In a program of quantitative character selection that was assessed or analyzed was what genetic variant was responsible for producing such character or quantitative traits that must be transmitted from parent to child in a predictable and reliable way [112]. That was, genetic variant predictions were produced in the form of ratios or indices among the emerging quantitative characters [36,50]. For example, to predict the inheritance of traits such as the number of egg production, the rate of growth, the graduation of life and the ability to convert food. In relation to the amount of egg production of a fish, one of the quantitative characteristics to be measured was the performance or index of gonad size and egg size [34,89]. Due to what genetic traits that produces or cause or control the development of the size of the gonads and fish eggs. Another thing was different from the rate of growth, then one of the quantitative characters that were seen or measured was the performance or index of the size of the stomach, the length and diameter of the intestine, the size of the mouth of the mouth, etc., because to cause or produce such traits there was a variety gene that controls or regulate it. For a live graduation selection process or sensitive to environmental conditions or the homeostatic abilities of a cultivated biota, one of the quantitative traits measured was the performance of the heart and other respiratory devices since there was a genetic variation that produces or controls it. While the character of the food conversion ability, the predicted quantitative trait was the digestive tools or channels involved in this digestive process, and all of this was controlled by a number of even the many different genes involved. This means the variety or variant of the size of the stomach, gonads, heart, intestine, and others was an expression of the genetic variation that controls and produces it. From the three variants or genetic variants of the additive variant, the dominant variant and the epistatic variant, only variants or additives could be transmitted in predictable and reliable ways, since the variety of additives was the function of the allele, while the dominant variety was a genotype function that was the interaction between an inter allele at each locus. So this variety of additives was sometimes regarded as a determinant factor in the breeding values of an individual [119,128]. This means that the value of performance or indexes used was a range which was considered as an additive variant. Whether the minimum or maximum distribution that will be used as a benchmark depends very much on the objectives to be achieved, e.g. whether, for ornamental fish or consumption fish, the mean value or even mode was the most frequent or frequent character trait could be used to predict or predict the genetic variants of the individual.
The individual deviations seen from the performance of cardiac indexes, stomach, meat, bone, body weight, body height, etc. could be used as additive variants which were further compared or converted to the size of the ideal or characteristic of a large and healthy individual could be used as a parameter for selection program [130,131]. Next look for the genetic distance and its cluster. As an example of the results displayed in the dendrogram, we could quickly sort through individuals that were the same or similar to other individuals based on the performance of the index was measured. In addition, it could be read how many degrees of similarity and differences in performance of phenotypic characters among individuals so that it could be considered in controlling or selecting individuals. Whether to continue or stop maintenance so as to save time, energy and production costs incurred [118,132]. Performance data indexes and criteria made could also be used to assess the performance index of the individual price when it was offered to the market so that the selling price may be different from the selling price of other individuals who have lower performance levels.
The value of the character performance index could also be used as a guide in assessing the existence or status of the population of this species in nature whether it still reaches the standard value of the index or not [133]. If an individual in a cultivated container exhibits a value greater than the value of this index, it may be assumed that the individual was superior and reliable, whereas if the value of the index was smaller, it could be assumed that the individual has already degraded or decreased performance so it needs anticipative steps in its management.
Engineering of aquaculture biota
Genetic engineering was essentially a set of techniques performed to manipulate genetic components, i.e., genomic DNA or genes that could be performed in a single cell or organism, even from one organism to another organism of a different kind [134,135]. In an effort to do genetic engineering, scientists use recombinant DNA technology. While organisms that were manipulated using recombinant DNA techniques were called genetically modified organisms or GMOs that have superior properties when compared to their original organisms [136,137]. Along with the advances in molecular biology today it allows scientists to take DNA from a species because DNA was easily extracted from cells. Then a molecular construction was constructed in the laboratory. DNA that has undergone molecular arrangement was called recombinant DNA while the gene isolated by the method was called a cloned gene.
Since the discovery of the structure of DNA by Watson and Crick in 1953, then began to develop genetic engineering technology in the 1970s with the aim to help create useful new products and organisms. History proves that genetic engineering constantly undergoes development and refinement of previous methods. Initially used conservative techniques pioneered by Gregor Mendel in the process of cross-breeding to get superior seeds that are hybrid [134,135]. This process takes a long time and has the disadvantage that was emerging properties that were not cool from the plant or animal parent. Until finally was born modern genetic engineering using recombinant DNA technology. Recombination was performed in vitro, so it was possible to modify specific genes and transfer them between different organisms such as bacteria, plants, and animals or could cloning only one desired gene in quick time. Since the onset of the development of genetic engineering, several techniques have been continuously improved and improved in order to lead to more advanced recombinant DNA technology [138-140]. The techniques that have been developed include polyploidization, androgenesis and gynogenesis, cloning, chimeras and transgenic.
Some of the steps involved in genetic engineering or recombinant DNA technology were as follows: DNA isolation containing target genes or genes of interest, isolation of bacterial DNA plasmids to be used as vectors, manipulation of DNA sequences through DNA insertion into vectors include: DNA cutting using endonuclease restriction enzymes and grafting into vectors using DNA ligase, transformation to host microorganism cells, cloning cells, identifying host cells containing recombinant DNA as desired and storage of cloned genes in DNA bank [139-141].
Genetic engineering has penetrated in many areas, not least the field of fisheries that produce superior quality fish, for example among others: zebrafish that were usually silver with purplish black stripes, after inserted with the jellyfish color gene injected into zebrafish could then red or green from his body [142,143]. The trigger gene from the jellyfish will activate the light beam on the fish when the fish was in an environment containing certain pollutant material.
Transgenic carp fish with growth reaches three times its normal size because it has a gene from growth hormone of rainbow trout that was transferred directly into carp fish eggs [144- 146]. Similarly, other studies have produced similar results, such as in red sea bream and Atlantic salmon which were also equally inserted by OPAFPcsGH gene growth hormone. Goldfish inserted with ocean pout antifreeze protein gene was expected to increase tolerance to cold weather [144]. Transgenic medaka fish capable of detecting mutations, especially those caused by pollutants, was very beneficial to the life of other aquatic animals and in the field of human health. The fish was after inserted with a mutagenic mutant vector and then the DNA vector was removed and inserted into the indicator bacterium that could calculate the mutant gene. Transgenic fish become durable and not quickly rot in storage after transplanted tomato genes [145]. But it could be otherwise if the application was intended for the world of agriculture, the fish genes that live in cold regions could be moved into the tomato to reduce the damage caused by freezing.
Induced breeding technique
Injecting was the act of entering the stimulating hormone into the female parent body. The commonly used stimulant hormone was ovaprim [146-149]. Way, catch the mature female parent gonad; suction 0.6 ml ovaprim for each kilogram of mains; injections of the parent's back; parent input that has been injected into another tub and leave for 10-12 h. Injections could also with a solution of the pituitary gland of goldfish. Way, catch the mature female parent gonad; prepare 2 kg of goldfish size 0.5 kg for each female parent; cut the goldfish vertically right behind the gills tutu; cut the head horizontally just below the eye; dispose of parts of the brain; take hypopisa glands; enter the pituitary gland into the crushing glass and crush it; input 1 cc aquabides and mix well; suction hypopisa solution; injections into the female parent's back; input the parent that has been injected into another tub and leave for 10-12 h [150-153].
Sperm picking was done half an hour before egg exposure. The trick, catch one male mature male; wipe to dry; parent body wrap with a small towel; massage into the genitalia; accommodating sperm into a plastic bowl or glass cup; mix 200 cc Sodium Chloride as physiological solution; stir until homogeneous. As note, sperm expenditure was done by two people. One person holding the head and massaging and another holding a plastic tail and bowl. Keep the sperm out of the water [150,152]. Egg discharge was done after 10-12 h after injection, but 9 h before checking. Procedure to remove eggs are prepare 3 pieces of plastic basin, a bottle of sodium chloride, a chicken feather, a duster and a tissue; catch the parent with the net screw; dry the mother body with a small towel or a towel; wrap the parent with a towel and leave the egg hole open; grasp the head by one person and grasp the tail by the other; massage the abdomen toward the hole by the head holder; eggs in a plastic basin; mix the sperm solution into the egg; mix well with chicken feathers; add sodium chloride and mix thoroughly; dispose of the liquid tCelcius degree lean the eggs from the blood; eggs ready [152,154].
Technical induced spawning
In induce spawning, eggs and sperm were not excreted, but the mother and the female were allowed to spawn on their own [154-156]. Spawning was done on the tub wall. The trick, prepare ready tub wall length 4 m, width 3 m and height 1 m; clean the mud and other dirt; dry for 3-4 days; water content as high as 80 cm; attach hapa to the same size as the tub; female injection at 06:00 AM; syringe back to the parent at 12:00 PM and feed into the spawning tub; injection of male parent at 12.00 PM and mix with the female parent; drain the water even more; let it spawn. Note: Spawning usually starts at 24.00 PM and ends in the morning.
Hatching grass carp fish eggs done in the aquarium. Procedures were prepare 20 pieces of aquarium size 60 cm long, 40 cm wide and 40 cm high; dry for 2 days; water content as high as 30 cm; install four aeration points for each aquarium and turn on during hatching; spread the stock evenly to the bottom surface of the aquarium; 2-3 days then remove some of the water and add new water until it reaches its original height [155,157]. Eggs will hatch in 2-3 days. Separating grass carp fish was done in a pool of land. Procedures were prepare pool size 500 m2; dry for 4-5 days; fix all parts of it; make channel of kemalir with width 40 cm and height 10 cm; level the ground; scatter 5-7 bags of chicken or quail dung; water content as high as 40 cm and soak for 5 days; stocking 50,000 larvae in the morning; after 2 days, give 1-2 kg of pelleted flour or pellet that has been soaked daily; seed harvesting after 3 weeks.
The second nursery was also done in a pool of soil. Procedures are prepare pool size 500 m2; dry 4-5 days; fix all parts of it; make channel with width 40 cm and height 10 cm; level the ground; scatter 5-7 bags of chicken or quail dung; water content as high as 40 cm and soak for 5 days, water not drained; stocking 40,000 seedlings from seedling I; give 2-4 kg of pelleted pellets or pellets daily; harvest the seeds done after a month old [154,155,158].
The third nursery was done in a pool of soil. How: prepare pool size 500 m2; dry 4-5 days; fix all parts of it; make its kemalir; level the ground; scatter 2 bags of chicken or quail dung; water content as high as 40 cm and soak for 5 days; stocking 30,000 heads from nursery II; give 4-6 kg of pellets; harvest the seeds done a month later. Enlargement of grass carp fish was done in the ground pool. How: prepare a pond size 500 m2; fix all parts of it; spread 6-8 bags of chicken or quail dung; water content as high as 40-60 cm and soak for 5 days; input of 10,000 seeds selected from nursery III; feed 3 percent daily, 3 kg at the start of maintenance and increase steadily in accordance with the weight of the fish; stream continuously; do the harvest after 2 months.
Sex reversal technique
Sex reversal technology was a genital transfer technique from female to male or vice versa, through hormonal administration and immersion techniques [159,160]. If the androgen hormones were given, the fish were directed to the male sex. But if given the hormone estrogen, sex was directed to a female. So, if the cultivators want to produce bullish, then the sex reversal process applied here uses the androgen hormones [161]. The androgen hormone used was 17-a methyltestosterone (C20H30O2). The white, powder-shaped hormone, produced by Sigma Chemical Co., Ltd., USA, but could be bought in chemicals stores, especially major cities in Indonesia. The amount of material required 20 ppm of solution of fish egg immersion. Each 300 fish eggs require 0.2 L of solution. How to make a soaking solution that dissolves 10 mg hormone methyltestosterone in 0.5 ml 70% alcohol, then diluted with distillate as much as 495 ml. This technology was used to get the super male parent or YY, which then produces children of fish with the sex of all males [162,163]. This technology is specific, so its application must be appropriate. Especially the type and dose of hormones, the duration of immersion, and the time to begin immersion. If the dose was less, then the sex of the fish will not change. But if the dose was excessive, it could cause the death of these fish. Even if not dead, the offspring tend to be sterile. A pond could produce fish consumption size of 125 g as much as 400-500 kg.
Gynogenesis technique
The purity of the carp mother must be returned. One way that could be done to restore the purity was to do inbreeding. But this method takes more than six generations. One generation takes 2 years, the time it takes to get the mother. So this way takes 12 years. To shorten the purification period could be done by gynogenesis. This method could change from 6 generations to 2 generations; pure strains could be obtained in the second generation. The success of this method depends on the thoroughness of the treatment and fertility of female gynogenesis.
Gynogenesis was the formation of zygote 2n or diploid without the genetic role of male gametes. So male gametes only function physically, so the process was only a parthenogenetic development of females or eggs. For that sperm irradiated. Radiation in gynogenesis aims to damage the spermatozoa chromosome so that at the time of conception does not function genetically. Spawning by way of gynogenesis will produce all male. Gynogenesis was a rare sexual reproduction in fertilization because the sperm nuclei that enter the egg in a state of inactivity so that the development of eggs was controlled by the female genetic only. Therefore, the descendants were replicas of the female parent both morphologically and in their genetic makeup. Artificial gynogenesis was performed through several treatments at the stage of conception and early embryonic development. This treatment aims to make the male genetic material inactive, and seek the occurrence of diploidization so that eggs could become a zygote. Genetic material in spermatozoa was made inactive by gamma-ray radiation, X-rays and ultraviolet light. Ultraviolet light was widely used because it was cheap.
The procedures of gynogenesis experiment were eggs originated from broodstock mains. In order to ovulate, the parent was injected with ovaprim or hypophysis extract gland. Sperm was taken from tawes fish as much as 1 ml, then diluted 100 times with a sodium chloride 0.9%. After diluted in radiation with ultraviolet light for 10 minutes. Eggs and sperm mixed, resulting in fertilization. After fertilization was dissolved in plastic sieves and soaked in water with a temperature of 25°C. After 2 min of fertilization in the heat shock at 40°C for 1.5-2 min. To remove the stickiness of eggs given tannin solution, after that incubated at a temperature of 28°C until hatching.
Androgenesis technique
The success of aquaculture, especially at the stage of enlargement one of them was determined by the quality of the seed. Because the seeds could live well, grow quickly and resistant to environmental changes and disease attacks. But goldfish seeds are good quality, hard to find in Indonesia. Because the quality of the parent has dropped considerably compared to twenty years ago.
Therefore genetics in fish should now be restored. One way of genetic improvement was by purifying the mother. One way that could be done was to do in-breeding. But this method takes more than six generations. One generation takes 2 years, the time it takes to get the mother. So this way takes 12 years. The practical way is to pass gynogenesis. In this way, the parent purification time could be shortened to six years. Another practical way was with androgenesis, a technology that utilizes the genetic properties of fish using biotechnology principles. This technique provides the possibility to speed up the purification time in fish selection. Androgenesis could be done by manipulating some fertilization process that was making the female genetic material gametes become inactive and seek to happen diploidization.
Female genetic material gametes could be made inactive with gamma-ray radiation, x-rays or ultraviolet light. Today ultraviolet light was more widely used because it was more practical and safer. Ultrasound radiation chould cause chromosomal damage. Based on the research of androgenesis by Arifin et al. [164], the result that radiation by using two TUV 15 watt lamps 30 cm away from the egg for 3-5 min has been able to non-act female gamete material. Surveillance was done to maintain embryo diploidy at an early stage of its development. Diploidization could be done by inhibiting the cleavage of mitosis I. The high degree of homozygosity could be achieved by surprise at the cleavage of mitosis I because the mitotic division of the resulting pair of chromosomes was identical from the paternal haploid genome that divides into two. Without the process of embryo diploidization resulting in the fertilization of non-active eggs will be haploid characterized abnormal.
The types of surprises that could be done include temperature shocks, using chemicals and electric shock. Temperature shock was one of the many methods to do because it was easy to apply explained that the use of temperature shocks was easier than the shock pressure. Heat shock has been commonly done to duplicate a set of chromosomes [165]. In the study of androgenesis of goldfish conducted, it was found that the time of heat shock made 40 min after fertilization at the best 40°C temperature was two minutes. Research on goldfish gynogenesis showed the diploid homozygous seed produced highest by heat shock 36-37 min after conception, states that generally the initial time of hot shock suppressing during cleavage mitosis I in gynogenesis was 40 could be done for 1.5-2.0 min [166-168].
The study of gynogenesis of goldfish by male parent tawes fish successfully produced gynogenetic seeds, with heat shock at 40°C after 40 min [169,170]. The highest gynogenetic production of carps fish obtained by administering a heat shock for one
Conclusion
Thus a study or review of the role and function of genetic analysis in the field of aquaculture. From the above discussions or studies, it could be concluded that: a true and profound understanding of the concept of genetics could provide a systematic and deep pattern of thought and analysis to be applied in the field of aquaculture; and application of genetic analysis that has been applied in the field of aquaculture, among others: observation of biota character performance, selection and breeding program and engineering of aquaculture biota.
References
- KKP (2018) Indonesian fisheries production statistics.
- FAO (2018) Fishery and Aquaculture Statistics. Global production by production source 1950-2016. FAO Fisheries and Aquaculture Department.
- Winsor E (1988) Mendelian genetics. Can Fam Physician 34: 859-862.
- Timmerman V, Strickland A, Buchner S (2014) Genetics of Charcot-Marie-Tooth (CMT) disease within the frame of the Human Genome Project success. Genes (Basel) 5: 13-32.
- Charlesworth B, Charlesworth D (2009) Darwin and genetics. Genetics 183: 757-766.
- Nguyen TTT, Hayes BJ, Ingram BA (2014) Genetic parameters and response to selection in blue mussel (Mytilus galloprovincialis) using a SNP-based pedigree. Aquaculture 420: 295-301.
- King RC, Stansfield WD, Mulligan PK (2006) A Dictionary of Genetics. 7th Edn, Oxford University Press.
- Kathiresan S, Srivastava D (2012) Genetics of human cardiovascular disease. Cell 148: 1242-1257.
- In VV, Connor WO, Dove M, Knibb W (2016) Can genetic diversity be maintained across multiple mass selection lines of Sydney rock oyster, Saccostrea glomerata despite loss within each? Aquaculture 454: 210-216.
- Lorenz C, Opitz R, Trubiroha A, Lutz I, Zikova A, et al. (2016) The synthetic gestagen levonorgestrel directly affects gene expression in thyroid and pituitary glands of Xenopus laevis tadpoles. Aquat Toxicol 177: 63-73.
- Duncan NJ, Sonesson AK, Chavanne H (2013) Principles of finfish broodstock management in aquaculture: Control of reproduction and genetic improvement. Advances in Aquaculture Hatchery Technology, pp: 23-75.
- Klug W, Cummings MP (1999) Essentials of genetics. Vasa.
- Saputra WA, Muslim AD, Sasanti (2014) The difference chromosomes number of snakehead fish (Channa striata) from floodplain area, highland swamp and tidal land. J Akuakultur Rawa Indones 2: 67-77.
- Gao YJ, Liu YJ, Chen XQ, Yang HJ, Li XF, et al. (2016) Effects of graded levels of histidine on growth performance, digested enzymes activities, erythrocyte osmotic fragility and hypoxia-tolerance of juvenile grass carp, Ctenopharyngodon idella. Aquaculture 452: 388-394.
- Lewandowski CM (2015) A brief mindfulness intervention on acute pain experience: an examination of individual difference. Exam Individ Differ 1: 1689-1699.
- Canada P (2018) Larval dietary protein complexity affects the regulation of muscle growth and the expression of DNA methyltransferases in Senegalese sole. Aquaculture 491: 28-38.
- Wang G (2010) Cloning and comparative studies of seaweed trehalose-6-phosphate synthase genes. Mar Drugs 8: 2065-2079.
- Petkeviciute E, Kania PW, Skovgaard A (2015) Genetic responses of the marine copepod Acartia tonsa (Dana) to heat shock and epibiont infestation. Aquat Rep 2: 10-16.
- Nolan MJ, Cribb TH (2005) The use and implications of ribosomal DNA sequencing for the discrimination of Digenean species. Adv Parasitol 60: 101-163.
- Jorgensen EH (2017) Biomarker response and hypothalamus-pituitary-internal axis functioning in Arctic charr from Bjornoya (74°30′ N), Norway, with high levels of organohalogenated compounds. Aquat Toxicol 187: 64-71.
- Furie K, Gwinn K (2009) Introduction to genetics. Stroke 40.
- Tsai HY (2017) Genotype imputation to improve the cost-efficiency of genomic selection in farmed Atlantic salmon. G3 (Bethesda) 7: 1377-1383.
- Yue GH, Wang L (2017) Current status of genome sequencing and its applications in aquaculture. Aquaculture 468: 337-347.
- Urgen BM (2018) Homozygous LAMC3 mutation links to structural and functional changes in visual attention networks. Neuroimage.
- Miller PA, Elliott NG, Vaillancourt RE, Kube PD, Koutoulis A (2014) Genetic diversity and pedigree assignment in tetraploid Pacific oysters (Crassostrea gigas). Aquaculture 433: 318-324.
- Felix MA (2012) Evolution in developmental phenotype space. Curr Opin Genet Dev 22: 593-599.
- Laimeheriwa BM (2014) Interaksi Faktor Lingkungan Dan Kontrol Hormonal Dalam Fisiologi Reproduksi Kepiting Bakau.
- Zamora AC, Montaldo HH, Montes GM, Rivas EG, Ortega AM, et al. (2015) Genetic parameters for body weight and survival in the Pacific white Shrimp Penaeus (Litopenaeus) vannamei affected by a White Spot Syndrome Virus (WSSV) natural outbreak. Aquaculture 447: 102-107.
- Hung D, Nguyen NH, Ponzoni RW, Hurwood DA, Mather PB (2013) Quantitative genetic parameter estimates for body and carcass traits in a cultured stock of giant freshwater prawn (Macrobrachium rosenbergii) selected for harvest weight in Vietnam. Aquaculture 404: 122-129.
- Lyu D, Wang W, Luan S, Hu Y, Kong J(2017) Estimating genetic parameters for growth traits with molecular relatedness in turbot (Scophthalmus maximus, Linnaeus) Aquaculture 468: 149-155.
- Guinand B (2017) Metapopulation patterns of additive and non-additive genetic variance in the sea bass (Dicentrarchus labrax). Ecol Evol 7.
- Olave M, Avila LJ, Sites JW, Morando M (2017) Hidden diversity within the lizard genus Liolaemus: Genetic vs. morphological divergence in the L. rothi complex (Squamata: Liolaeminae). Mol Phylogenet Evol 107: 56-63.
- Li Y, Li W, Xie D, Wang Y, Cheung EFC, et al. (2018) Grey matter reduction in the caudate nucleus in patients with persistent negative symptoms: An ALE meta-analysis. Schizophr Res 192: 9-15.
- Nolasco-Alzaga HR, Perez-Enriquez R, Enez F, Bestin A, Mechetnov EP, et al. (2018) Quantitative genetic parameters of growth and fatty acid content in the hemolymph of the White leg shrimp Litopenaeus vannamei. Aquaculture 482: 17-23.
- Zhang G (2018) Construction of high-density genetic linkage maps and QTL mapping in the golden pompano. Aquaculture 482: 90-95.
- Laimeheriwa BM, As K, Yl N, Gv L (2018) The role of biomarker as a taxonomic material and indicator of characters performance on marine biota. J Mar Sci 8.
- Liu JJ (2018) Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J Dairy Sci 101: 433-444.
- Evans DM, Gillespie NA, Martin NG (2002) Biometrical genetics. Biol Psychol 61: 33-51.
- Birchler JA (2015) Mendel, mechanism, models, marketing and more. Cell 163: 9-11.
- Gayon J (2016) From Mendel to epigenetics: History of genetics. C R Biol 339: 225-230.
- Neher RA, Shraiman BI (2011) Statistical genetics and evolution of quantitative traits. Rev Mod Phys 83.
- Li S, Cai W, Zhou B (1993) Variation in morphology and biochemical genetic markers among populations of blunt snout bream (Megalobrama amblycephala). Aquaculture 11: 117-127.
- Barros J, Velasco LA, Winkler FM(2018)Heritability, genetic correlations and genotype by environment interactions in productive traits of the Caribbean scallop, Argopecten nucleus (Mollusca: Bivalvia). Aquaculture 488: 39-48.
- Macbeth M, Kenway M, Salmon M, Benzie J, Knibb W, et al. (2007) Heritability of reproductive traits and genetic correlations with growth in the black tiger prawn Penaeus monodon reared in tanks. Aquaculture 270: 51-56.
- Acquaah G (2017) Plant Breeding Principles. B. Thomas, B.G. Murray and D.J.B.T.E. of A. P. Oxford: Academic Press, pp: 236-242.
- Kenway M (2006) Heritability and genetic correlations of growth and survival in black tiger prawn Penaeus monodon reared in tanks. Aquaculture 259: 138-145.
- Xiong XM, Chen YL, Liu LF, Wang W, Robinson NA, et al. (2017) Estimation of genetic parameters for resistance to Aeromonas hydrophila in blunt snout bream (Megalobrama amblycephala). Aquaculture 479: 768-773.
- Liu J (2015) Genetic parameters and selection responses for growth and survival of the small abalone Haliotis diversicolor after four generations of successive selection. Aquacultuer 436: 58-64.
- JMarotti JD, Schnitt SJ (2018) Genotype-phenotype correlations in breast cancer. Surg Pathol Clin 11: 199-211.
- Laimeheriwa BM (2017) Phenetic relationship study of gold ring cowry, Cypraea Annulus (Gastropods: Cypraeidae) in Mollucas Islands based on shell morphological. Fish Aquat J 8.
- Laimeheriwa BM, Khouw AS, Natan YL, Limmon GV (2018) Phenotypic classification of molluscs ring cowrie, Cypraea annulus Linn., 1758 (gastropod: cypraeidae), based on shell meristical. J Aquacult Mar Biol 1758: 80-90.
- Miller AJ, Roman B, Norstrom E (2016) A method for easily customizable gradient gel electrophoresis. Anal Biochem 509: 12-14.
- Tang L, Wang CX, Shu-Lin Liu SS (2017) Pulsed field gel electrophoresis. Elsevier.
- Hachmoller O (2017)Investigating the influence of standard staining procedures on the copper distribution and concentration in Wilson’s disease liver samples by laser ablation-inductively coupled plasma-mass spectrometry. J Trace Elem Med Biol 44: 71-75.
- Costa PM (2018) Chapter 4 - Staining Protocols. P. M. B. T.-T. H. of H. P. in A. E. Costa, Ed. Academic Press, pp: 83-117.
- Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248-249.
- Osman MI (1997) Exploring a mixture of distributions using minitab. Comput Biol Med 27: 223-232.
- Delozier MR, Orlich S (2005) Discovering influential cases in linear regression with MINITAB: Peeking into multidimensions with a MINITAB macro. Stat Methodol 2: 71-81.
- Liu RX, Kuang J, Gong Q, Hou XL (2003) Principal component regression analysis with SPSS. Comput Methods Programs Biomed 71: 141-147.
- Lees MM (1997) Current genetics. Nurse Pract 22: 153.
- Zhou W, Geiersbach K, Chadwick B (2017) Rapid removal of cytology slide coverslips for DNA and RNA isolation. J Am Soc Cytopathol 6: 24-27.
- Porta JM, Porta J, Alvarez MC (2007) Development of a PCR multiplex system in the sea bass (Dicentrarchus labrax) and its application in the population genetic studies. Aquaculture 272: 300-301.
- Higgins D, Sharp PM (1988) CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73: 237-244.
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23(10): 403-405.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874.
- Yeung JWY, Zhou GJ, Leung KMY (2017) Spatiotemporal variations in metal accumulation, RNA/DNA ratio and energy reserve in Perna viridis transplanted along a marine pollution gradient in Hong Kong. Mar Pollut Bull 124: 736-742.
- Yandi I, Altinok I (2018) Irreversible starvation using RNA/DNA on lab-grown larval anchovy, Engraulis encrasicolus and evaluating starvation in the field-caught larval cohort. Fish Res 201: 32-37.
- Alam MS, Islam MS (2005) Population genetic structure of Catla catla (Hamilton) revealed by microsatellite DNA markers. Aquaculture 246: 151-160.
- Mi X (2015) Potential DNA markers as a rapid tracing tool for animal adulterants in vegetarian food. Food Res Intvol 76: 926-931.
- Anshary H (2011) Molecular Identification with PCR-RFLP technique parasitic larvae Anisakis spp. (Nematodes: Anisakidae) on Tongkol fish (Auxis thazard) and Kembung (Restrelliger kanagurta) from Makassar waters. 13: 70-77.
- Cui C (2017) Determination of genetic diversity among Saccharina germplasm using ISSR and RAPD markers. C R Biol 340: 76-86.
- Patel P, Rajkumar BK, Parmar P, Shah R, Krishnamurthy R (2017) Assessment of genetic diversity in Colletotrichum falcatum Went accessions based on RAPD and ISSR markers. J Genet Eng Biotechnol.
- Ayuningrum PI, Afrianto E, Mulyani Y (2012) Genetic diversity of seaweed Eucheuma spp. from Sukabumi, West Java based on PCR RAPD method. J Fish Mar 3: 337-345.
- Kong L, Bai J, Li Q (2014) Comparative assessment of genomic SSR, EST-SSR and EST-SNP markers for evaluation of the genetic diversity of wild and cultured Pacific oyster, Crassostrea gigas Thunberg. Aquaculture 420: 85-91.
- Liu F, Yao J, Wang X, Repnikova A, Galanin DA, et al. (2012)Genetic diversity and structure within and between wild and cultivated Saccharina japonica (Laminariales, Phaeophyta) revealed by SSR markers. Aquaculture 358:139-145.
- Pan L (2017) Genetic diversity among germplasms of Pitaya based on SSR markers. Sci Horticult (Amsterdam) 225: 171-176.
- Zhao J, Han D, Shi K, Wang L, Gao J, et al. (2018) Influence of epistatic segregation distortion loci on genetic marker linkages in Japanese flounder. Genomics 110: 59-66.
- Ning Y, Liu X, Wang ZY, Guo W, Li Y, et al. (2007) A genetic map of large yellow croaker Pseudosciaena crocea. Aquaculture 264: 16-26.
- Gong C (2013) Identification and phylogenetic analysis of a CC-NBS-LRR encoding gene assigned on chromosome 7B of wheat. Int J Mol Sci 14: 15330-15347.
- Xie XH, Yu ZG, Han GS, Yang WF, Anh V (2015) Whole-proteome based phylogenetic tree construction with inter-amino-acid distances and the conditional geometric distribution profiles. Mol Phylogenet Evol 89: 37-45.
- Lewin AC (2018) Genomic, recombinational and phylogenetic characterization of global feline Herpes virus 1 isolates. Virology 518: 385-397.
- Ruzgar E, Erciyes K (2012) Clustering based distributed phylogenetic tree construction. Expert Syst Appl 39: 89-98.
- Al-Ruqaie I, Al-Khalifah NS, Shanavaskhan AE (2016) Morphological cladistic analysis of eight popular Olive (Olea europaea L.) cultivars grown in Saudi Arabia using numerical taxonomic system for personal computer to detect phyletic relationship and their proximate fruit composition. Saudi J Biol Sci 23: 115-121.
- Kalinowski T, Taper ST, Marshall ML (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16: 1099-1106.
- Lincoln S, Daly M, Lander E (1992) Mapping genes controlling quantitative traits with MAPMAKER/QTL 1.1. 2nd edn. Whitehead Institute Technical Report.
- Faqih I, Soelistyowati D, Carman O (2015) Phenotypes performance of tilapia best, nirwana II, jatimbulan, and sultana using floating net and pond culture system. J Iktiologi Indones 15: 193-200.
- Vandeputte M (2016) Quantitative genetic variation for post-stress cortisol and swimming performance in growth-selected and control populations of European sea bass (Dicentrarchus labrax). Aquaculture 455: 1-7.
- Henriksson PJG, Dickson M, Allah AN, Al-Kenawy D, Phillips M (2017) Benchmarking the environmental performance of best management practice and genetic improvements in Egyptian aquaculture using life cycle assessment. Aquaculture 468: 53-59.
- Silva PF, McGurk C, Thompson KD, Jayasuriya NS, Bron JE (2015) Development of a quantitative semi-automated system for intestinal morphology assessment in Atlantic salmon, using image analysis. Aquaculture 442: 100-111.
- Qin Y, Liu X, Zhang H, Zhang G, Guo X (2007) Genetic mapping of size-related quantitative trait loci (QTL) in the bay scallop (Argopecten irradians) using AFLP and microsatellite markers. Aquaculture 272: 281-290.
- Chen W, Xia X, Huang Y, Chen X, Han JDJ (2016) Bioimaging for quantitative phenotype analysis. Methods 102: 20-25.
- Huang D, Walla JA, Dai W (2014) Quantitative phenotyping of X-disease resistance in chokecherry using real-time PCR. J Microbiol Methods 98: 1-7.
- Wendte JM, Pikaard CS (2017) The RNAs of RNA-directed DNA methylation. Biochim Biophys Acta Gene Regul Mech 1860: 140-148.
- Fellahi S (2017) Comparison of two techniques of adiponectin assay, ELISA and immunoturbidimetry: Should we move towards standardization? Diabetes Metab 43: 395-397.
- Terato K, Do C, Chang J, Waritani T (2016) Preventing further misuse of the ELISA technique and misinterpretation of serological antibody assay data. Vaccine 34: 4643-4644.
- Samsonova JV, Osipov AP, Kondakov SE (2017) Strip-dried whole milk sampling technique for progesterone detection in cows by ELISA. Talanta 175: 143-149.
- Hernandez SIDLC (2018) Real-time RT-PCR, a necessary tool to support the diagnosis and surveillance of rotavirus in Mexico. Diagn Microbiol Infect Dis 90: 272-276.
- Dang-Trinh MA (2018) Utilization of real time PCR for the assessment of egg burden in the organs of Schistosoma japonicum experimentally infected mice. Exp Parasitol 189: 61-65.
- Pranawaty RN, Buwono dan E, Liviawaty ID (2012) Applications of conventional polymerase chain reaction (PCR) and real-time PCR for detection of white spot syndrome virus on crabs. J Perikan dan Kelaut 3: 61-74.
- Macbeth M, Kenway M, Salmon M, Benzie J, Knibb W, et al. (2007) Heritability of reproductive traits and genetic correlations with growth in the black tiger prawn Penaeus monodon reared in tanks. Aquaculture 270: 51-56.
- Vieira VLA, Norris A, Johnston (2007) Heritability of fibre number and size parameters and their genetic relationship to flesh quality traits in Atlantic salmon (Salmo salar L.). Aquaculture 272: 100-109.
- Laconisi V (2017) Dietary inclusion of Tenebrio molitor larvae meal: Effects on growth performance and final quality treats of blackspot sea bream (Pagellus bogaraveo). Aquaculture 476: 49-58.
- Barros J, Velasco LA, Winkler FM (2018) Heritability, genetic correlations and genotype by environment interactions in productive traits of the Caribbean scallop, Argopecten nucleus (Mollusca: Bivalvia). Aquaculture 488: 39-48.
- Farias WJ, Winkler FM, Brokordt KB (2017) Genotype by environment interactions, heritabilities and genetic correlations for productive traits of Haliotis rufescens. Aquacult 473: 407-416.
- Rye M, Lillevik KM (1990) Survival in the early fresh-water period in Atlantic salmon (Salmo salar) and rainbow trout (Salmo gairdneri): Heritabilities for survival and genetic correlation between survival and growth. Aquaculture 85: 328-329.
- Hillen JEJ (2017) Estimates of genetic variability and inbreeding in experimentally selected populations of European sea bass. Aquaculture 479: 742-749.
- Winkelman A, Peterson RG (1994) Genetic parameters (heritabilities, dominance ratios and genetic correlations) for body weight and length of Chinook salmon after 9 and 22 months of saltwater rearing. Aquaculture 125: 31-36.
- Garcia-Celdran M, Ramis G, Manchado M, Estevez A, Navarro A, et al.(2015)Estimates of heritabilities and genetic correlations of raw flesh quality traits in a reared gilthead sea bream (Sparus aurata L.) population sourced from broodstocks along the Spanish coasts. Aquaculture 446: 181-186.
- Domingos JA, Smith-Keune C, Robinson N, Loughnan S, Harrison P, et al. (2013) Heritability of harvest growth traits and genotype-environment interactions in barramundi, Lates calcarifer (Bloch). Aquaculture 402-403: 66-75.
- Garcia-Celdran M (2015) Estimates of heritabilities and genetic correlations of growth and external skeletal deformities at different ages in a reared gilthead sea bream (Sparus aurata L.) population sourced from three broodstocks along the Spanish coasts. Aquaculture 445: 33-41.
- Bentsen HB (2017) Genetic improvement of farmed tilapias: Response to five generations of selection for increased body weight at harvest in Oreochromis niloticus and the further impact of the project. Aquaculture 468: 206-217.
- Chavanne H (2016) A comprehensive survey on selective breeding programs and seed market in the European aquaculture fish industry. Aquacult Int 24(5).
- Thodesen J, Rye M, Wang YX, Bentsen HB, Gjedrem T (2012) Genetic improvement of tilapias in China: Genetic parameters and selection responses in fillet traits of Nile tilapia (Oreochromis niloticus) after six generations of multi-trait selection for growth and fillet yield. Aquaculture 366: 67-75.
- Saura M, Villanueva B, Fernandez J, Toro MA (2017) Effect of assortative mating on genetic gain and inbreeding in aquaculture selective breeding programs. Aquaculture 472: 30-37.
- Palti Y, Rexroad CE, Welch T, Wiens G, Silverstein J (2007) Selective breeding and genetic mapping of disease resistance in rainbow trout. Aquaculture 272.
- Correa K, Bangera R, Figueroa R, Lhorente JP (2017) The use of genomic information increases the accuracy of breeding value predictions for sea louse (Caligus rogercresseyi) resistance in Atlantic salmon (Salmo salar). Genet Sel Evol 49.
- Kause A, Ritola O, Paananen T, Wahlroos H, Mantysaari EA (2005) Genetic trends in growth, sexual maturity and skeletal deformations and rate of inbreeding in a breeding programme for rainbow trout (Oncorhynchus mykiss). Aquaculture 247: 177-187.
- Nino EL, Cameron Jasper W (2015) Improving the future of honey bee breeding programs by employing recent scientific advances. Curr Opin Insect Sci 10: 163-169.
- Janssen K, Chavanne, Berentsen P, Komen H (2017) Impact of selective breeding on European aquaculture. Aquaculture 472.
- Domingos JA, Smith-Keune C, Harrison P, Jerry DR (2014) Early prediction of long-term family growth performance based on cellular processes — A tool to expedite the establishment of superior foundation broodstock in breeding programs. Aquaculture, pp: 88-96.
- Caligari PDS, Brown J (2017) Plant breeding. Oxford: Academic Press, pp: 229-235.
- Difford GF, Vlok AC, Rhode C, Brink D (2017) Heritability of growth traits in South African Abalone (Haliotis midae L.) using the ‘internal reference’ method. Aquaculture 468: 451-457.
- Murray BG (2017) Hybridization and plant breeding. Oxford: Academic Press, pp: 168-173.
- Kettunen A, Fjalestad KT (2007) Genetic parameters for important traits in the breeding program for Atlantic cod (Gadus morhua L.). Aquacult 272.
- Domingos JA, Smith-Keune C, Jerry DR (2014) Fate of genetic diversity within and between generations and implications for DNA parentage analysis in selective breeding of mass spawners: A case study of commercially farmed barramundi, Lates calcarifer. Aquaculture 424: 174-182.
- Aljumaah RS (2018) Simulated genetic gain of a close breeding program for Ardi goat in Saudi Arabia. J Saudi Soc Agric Sci.
- Loughnan SR, Smith-Keune C, Jerry DR, Beheregaray LB, Robinson NA (2016) Genetic diversity and relatedness estimates for captive barramundi (Lates calcarifer, Bloch) broodstock informs efforts to form a base population for selective breeding. Aquacult Res 47.
- de Lago A, Rezende TT, Dias MAD, de Freitas RTF, Hilsdorf AWS (2017) The development of genetically improved red tilapia lines through the backcross breeding of two Oreochromis niloticus strains. Aquacult 472: 17-22.
- Skaarud A, Woolliams JA, Gjoen HM (2014) Optimising resources and management of genetic variation in fish-breeding schemes with multiple traits. Aquaculture 420: 133-138.
- Ritola O, Paananen T, Kause A (2007) Successful divergent selection for growth and age at maturity in the Finnish breeding programme for rainbow trout: Genetic responses in two selection lines. Aquaculture 272.
- Janssen K, Chavanne H, Berentsen P, Komen H (2017) Impact of selective breeding on European aquaculture. Aquaculture 472.
- Liu J (2015) Genetic parameters and selection responses for growth and survival of the small abalone Haliotis diversicolor after four generations of successive selection. Aquaculture 436:58-64.
- de Araujo EP (2017) Dietary spray-dried plasma enhances the growth performance, villus: crypt ratio and cold-induced stress resistance in Nile tilapia (Oreochromis niloticus). Aquaculture 479: 675-681.
- Myhr AI, Dalmo RA (2005) Introduction of genetic engineering in aquaculture: Ecological and ethical implications for science and governance. Aquaculture 250: 542-554.
- Ushasree MV, Shyam K, Vidya J, Pandey A (2017) Microbial phytase: Impact of advances in genetic engineering in revolutionizing its properties and applications. Bioresour Technol 245: 1790-1799.
- Wardowski TT, Małyska A (2015)Uninformed and disinformed society and the GMO market. Trends Biotechnol 33: 1-3.
- Fraiture MA (2017) An integrated strategy combining DNA walking and NGS to detect GMOs. Food Chem 232: 351-358.
- Clark DP, Pazdernik NJ (2016) Recombinant DNA technology. Eds. Boston. Academic, pp: 63-95.
- Hacker D, Wurm FM (2017) Recombinant DNA technology for production of protein therapeutics in cultured mammalian cells. Module in Life Sciences Elsevier.
- Oliveira C, Aguiar TQ, Domingues L (2017) Principles of genetic engineering current developments in biotechnology and bioengineering. Elsevier, pp: 81-127.
- Carter M, Shieh J (2015) Molecular cloning and recombinant DNA technology. Guide to Research Techniques in Neuroscience (2nd Edn) Eds. San Diego: Academic Press, pp: 219-237.
- Powers DA, Chen TT (1995) Genetic engineering to improve the growth characteristics of finfish and shellfish for aquaculture in Pacific Rim countries: Cloning, expression, and gene transfer of growth hormone and insulin-like growth factors. Aquaculture 135.
- Prather RS, Whitworth KM, Schommer SK, Wells KD (2017) Genetic engineering alveolar macrophages for host resistance to PRRSV. Vet Microbiol 209: 124-129.
- Liu Y, Zhang W, Yong L, Zhao X, Jia X, et al. (2011) An Assessment of androgenic/anti-androgenic effects of GH Transgenic carp by Hershberger assay. Biomed Environ Sci 24: 445-449.
- Zhong C (2013) Increased food intake in growth hormone-transgenic common carp (Cyprinus carpio L.) may be mediated by upregulating Agouti-related protein (AgRP). Gen Comp Endocrinol 192: 81-88.
- Kurdianto, Alimuddin, Faridah N, Yoshizaki G, Nuryati S, et al. (2016) Growth, survival and body composition of transgenic common carp Cyprinus carpio 3rd generation expressing tilapia growth hormone cDNA. Hayati J Biosci 23: 150-154.
- Shaughnessy PJO (2018) Follicle stimulating hormone (FSH) and male reproduction in mammals. Module in Biomedical Sciences. Elsevier.
- De Pascali F (2018) Follicle-stimulating hormone receptor: Advances and remaining challenges. Academic Press.
- Pouwer AW, Farquhar C, Kremer JAM, Marjoribanks J (2016) Long-acting follicle-stimulating hormone versus daily follicle-stimulating hormone for women undergoing assisted reproduction. Fertil Steril 105: 1454-1456.
- Zadmajid V, Mirzaee R, Hoseinpour H, Vahedi N, Butts IAE (2017) Hormonal induction of ovulation using OvaprimTM [(D-Arg6, Pro9NEt)-sGnRH+domperidone] and its impact on embryonic development of wild-caught Longspine scraper, Capoeta trutta (Heckel), 1843. Anim Reprod Sci 187: 79-90.
- Zadmajid V (2016) Comparative effects of human chorionic gonadotropin (hCG) and OvaprimTM (sGnRHa+domperidone) on the reproductive characteristics of wild-caught male Longspine scraper, Capoeta trutta (Heckel, 1843). Aquaculture 463: 7-15.
- Das P (2016) Comparative efficacy of different inducing agents on breeding performance of a near threatened cyprinid Osteobrama belangeri in captivity. Aquacult Rep 4: 178-182.
- Targonska K, Kucharczyk D, Kujawa R, Mamcarz A, Zarski D (2010) Controlled reproduction of asp, Aspius aspius (L.) using luteinizing hormone releasing hormone (LHRH) analogues with dopamine inhibitors. Aquaculture 306: 407-410.
- DiMaggio MA, Broach JS, Ohs CL (2013) Evaluation of ovaprim and human chorionic gonadotropin doses on spawning induction and egg and larval quality of pinfish, Lagodon rhomboids. Aquaculture 414-415: 9-18.
- Dong S (2017) Channel catfish Ictalurus punctatus strain comparison for induced ovulation in the early spawning season to produce channel catfish ♀×blue catfish I. furcatus ♂ hybrid catfish embryos. Aquaculture 471: 185-189.
- Flajshans M, Kvasnicka P, Rab P (1993) Genetic studies in tench (Tinca tinca L.): High incidence of spontaneous triploidy. Aquacult 110: 243-248.
- Harith H, Husain ML, Akhir MFM (2016) Coastal oceanographic processes associated with blood cockle (Anadara granosa) induce spawning season in Kapar, Selangor, Malaysia. J Ocean Eng Sci 1: 289-299.
- Trá»ÂÂng TQ, van Arendonk JAM, Komen H (2013) Genetic parameters for reproductive traits in female Nile tilapia (Oreochromis niloticus): I. Spawning success and time to spawn. Aquaculture 416: 57-64.
- Ferguson-Smith MA (2013) Sex reversal. Brenner’s Encyclopedia of Genetics (2nd Edn), Academic Press, pp: 414-415.
- Fatima S, Adams M, Wilkinson R (2016) Sex reversal of brook trout (Salvelinus fontinalis) by 17α-methyltestosterone exposure: A serial experimental approach to determine optimal timing and delivery regimes. Anim Reprod Sci 175: 39-47.
- Horie Y, Myosho T, Sato T, Sakaizumi M, Hamaguchi S, et al. (2016) Androgen induces gonadal soma-derived factor, Gsdf, in XX gonads correlated to sex-reversal but not Dmrt1 directly, in the teleost fish, northern medaka (Oryzias sakaizumii).Mol Cell Endocrinol 436: 141-149.
- Luckenbach JA, Fairgrieve WT, Hayman ES(2017)Establishment of monosex female production of sablefish (Anoplopoma fimbria) through direct and indirect sex control. Aquaculture 479: 285-296.
- Ayuningtyas SQ, Junior MZ, Soelistyowati DT (2015) Sex reversal of red tilapia using 17α-methyltestosterone-enriched feed and increased temperature. J Akuakultur Indones 14: 159-163.
- Arifin OZ, Subagja J, Hadie W (2015) Biometric characterization three population of semah mahseer Tor douronensis (Valenciennes, 1842) in support to conservation of genetic resources. JIktiologi Indones 15: 143-154.
- Hidayat A, Carman O (2016) Sexual dimorphism related to growth in climbing perch Anabas testudineus. J Akuakultur Indones 15: 8-14.
- Sun Y, Zhang C, Liu S, Duan W, Liu Y (2007) Induced interspecific androgenesis using diploid sperm from allotetraploid hybrids of common carp × red crucian carp. Aquaculture 264: 47-53.
- Hatef A (2012) Anti-androgen vinclozolin impairs sperm quality and steroidogenesis in goldfish. Aquat Toxicol 122: 181-187.
- Depince A, Marandel L, Goardon L, Le Bail PY, Labbe C (2011) Trout coelomic fluid suitability as Goldfish oocyte extender can be determined by a simple turbidity test. Theriogenology 75: 1755-1761.
- Linhart O (1995) Genetic studies with tench, Tinca tinca L.: Induced meiotic gynogenesis and sex reversal. Aquaculture 132: 239-251.
- Lahrech Z, Kishioka C, Morishima K, Mori T, Saito S, et al. (2007) Genetic verification of induced gynogenesis and microsatellite-centromere mapping in the barfin flounder, Verasper moseri. Aquaculture 272: 115-124.
- Komen H, Thorgaard GH (2007)Androgenesis, gynogenesis and the production of clones in fishes: A review. Aquaculture 269:150-173.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences