ISSN : 2573-4466
Insights in Enzyme Research
Carboxylesterases: Sources, Characterization and Broader Applications
Samta Sood1, Abhishek Sharma2, Nikhil Sharma1 and Shamsher Singh Kanwar2
1 Sub Distributed Information Centre, Himachal Pradesh University, Summer Hill, Shimla, India
2 Department of Biotechnology, Himachal Pradesh University, Summer Hill, Shimla, India
- *Corresponding Author:
- Shamsher Singh Kanwar
Department of Biotechnology, Himachal Pradesh University, Summer Hill, Shimla-171 005, India.
Tel: +911772831948
Fax: +911772832153
E-mail: kanwarss2000@yahoo.com
Received Date: November 20, 2016; Accepted Date: December 09, 2016; Published Date: December 12, 2016
Citation: Sood S, Sharma A, Sharma N, et al. Carboxylesterases: Sources, Characterization and Broader Applications. Insights Enzyme Res. 2016, 1:1.doi: 10.21767/2573-4466.100002
Abstract
Carboxylesterases (CEs) are a group of versatile lipolytic enzymes capable of catalyzing the hydrolysis of esters into acid and alcohol molecules. These enzymes are extensively used in diverse xenobiotic and endobiotic degradations, biocatalysis, and drug metabolism. The present review article focuses on structure, function and major applications of CEs mainly sourced from bacteria and archaea. The CEs are divided in different families (15 families) depending upon their source, biochemical properties, common pentapeptide motif (GXSXG) with catalytic Ser, an entirely different GDSL motif, (family II)/ SXXK motif (family VIII), position of catalytic triad and their protein forms. CEs find diverse applications in degradation of xenobiotic compounds, biocatalysis, biotransformation of compounds such as cholesterol, synthesis of optically active compounds, food industry, anticancer therapeutics, drug and prodrugs like aspirin, delapril etc.
Keywords
Carboxylesterases (CEs); Structure; Function; Families; Mechanism; Properties; Distribution
Introduction
Esterases, particularly carboxylesterases (CEs) belong to the family of serine esterases, which hydrolyze simple esters (ethyl acetate) and triglycerides of short chain length (C ≤ 6) [1]. These enzymes catalyze the hydrolysis of short-chain aliphatic as well as aromatic carboxylic ester compounds and are often inhibited by small concentration of organophosphate (OP) compounds [2]. Esterase-catalyzed reactions are mostly specific for an alcohol or acid moiety of the substrate but not for both. A carboxylesterase from Pyrobacculum calidifontis VA-1 has a strong affinity for an alcohol moiety [3,4]. Structurally and functionally these enzymes are similar to the other members of α/β hydrolase fold superfamily, but have less amino acid sequence identity with other members of this family [5]. However, these enzymes are multifunctional in nature. A single estereolytic reaction can catalyze the hydrolysis of esters as well as amides. As in signature amidases which contain conserved amino acid sequence of 130 amino acids rich in Gly (G) and Ser (S) but lack Asp (D) and His (H) that are typical characteristic of carboxylesterases. The substrate affinity for substrates like ethyl acetate, ethyl butyrate, tributyrin as well as phenyl acetate classify carboxyl ester hydrolases into acetylesterases, arylesterases and carboxylesterases. The substrate specificity of CEs also divides them into two categories; specific and nonspecific. Being adapted to different functions some CEs have strong substrate selectivity (e.g., acetylcholinesterases) whereas others that are less selective and are referred as true esterases [5,6]. Carboxylesterases have broad substrate specificity due to large conformable active site, which allows structurally diverse substrates to sit at it. The CEs have been classified on the basis of sequence similarity into eight families [7] but later on classification extended and their number increased. Their broad substrate specificity enables them to be involved in the evolution of carbon sources and diverse catabolic pathways [8]. Many people classified them differently based on their insecticide resistance into EstA, EstB and EstC [9]. Because of their inhibition by organophosphate compounds (OP’s), CEs are defined as EstB. The chromophoric substances e.g., paranitrophenyl esters or tributyrin-supplemented agar plates are commonly used for the screening of microbial CEs. Active esterase-producing organisms form clear halos in agar Petri plates containing soluble substrates such as triglycerides. Hydrolysis of these dispersed lipids results in the formation of clearing zones around the bacterial colonies [10]. Enzyme assays to determine the esterase activity include spectrophotometric and saponification methods [11]. These enzymes help to metabolize drug esters and amides carbamates [12].
Carboxylesterases sources
CEs have been isolated from diverse sources including bacteria, fungi, algae, animals, plants and human beings. A number of carboxylesterase genes have been cloned and expressed in suitable host systems. They are present in all environments including moderate and extreme temperature range habitats which shape these enzymes. CEs have also been reported from cold- and salt-resistant marine microorganisms. The hyperthermophilic microbial sources mostly include Archaebacteria. The first CE was reported from a thermo-acidophilic bacterium Sulfolobus acidocaldarius and its 3D structure was also elucidated. Another, esterase elucidated from genus Alkalibacterium sp. was the first reported esterase from halophilic cold-adapted environment [13].
Metagenomic sources
Metagenomic samples are quite commonly used for the isolation of CEs. Most of the esterases have been isolated by metagenomics approach which is a cultureless method [14,15]. It is a common technique to explore novel enzymes that have potential for industrial applications. The EstCS2 isolated from metagenomic samples can hydrolyze (R/S-ketoprofen ethylester) with great enantioselectivity for R-enantiomer of it. Ketoprofenethylester is a non-steroidal anti-inflammatory drug (NSAID) having analgesic, antipyretic and anti-inflammatory affects as well as great enantioselectivity for R-enantiomer [16]. Insect-derived CEs are categorized into α-esterases, β-esterases, juvenile hormone esterase, gleotactins, neurotactins, neuroligins and glutactin classes.
Bacterial sources
Because of the ease with which bacterial cells may be mass cultured and genetically manipulated, the bacterial strains are considered superior sources of these enzymes than the higher organisms. However, CEs have also been isolated from the higher animals like mammals. The genomes from most hyperthermophilic archaeons have been well characterized and have been completely sequenced too, e.g., Archaeoglobus fulgidus [17].
Human carboxylesterases
In human, CEs (hCEs) these are present in the form of isozymes. They are present in different organs in human body like brain, liver, kidneys, muscles and adipose tissue. Within a cell, the carboxylesterases are confined to endoplasmic reticulum. Human genome organization database listed five types of carboxylesterase genes family CES1, CES2, CES3, CES4, and CES5 [18]. An additional CES6 gene family also includes structures of many mammalian carboxylesterase [19]. In mammals, the first crystal structure was deciphered for rabbit liver carboxylesterase (rCE). Its structure bears 81% sequence identity with hCE-1.
General Structure of CEs
The amino acid sequence of CEs belongs to superfamily of α/β hydrolase enzymes. The structure of the enzyme consists of central eight stranded β-sheets surrounded by helices and connecting loops. The catalytic structure of these enzymes is made up of Ser (S), His (H) and Asp (D). Ser (S) is generally found fixed in a pentapeptide motif (GXSXG) where X is any residue. Nucleophile elbow is a sharp turn present in the 3D structure and is produced by this motif. This nucleophilic elbow is located at the apex of sharp turn between α-helix and β-sheet. The structure of CE enzyme was first determined from Pseudomonas fluorescens. Use of serine protease inhibitor, phenylmethylsulphonyl fluoride (PMSF) and isomorphous replacement method revealed the CEcatalytic triad comprising Ser-114, His-199 and Asp-168 for first time within the structure of Pseudomonas fluorescens. An open active site and a large binding pocket for acid moiety of substrate contribute towards the broad substrate specificity of CE [20]. The backbone carbonyl of the residue was located three residues upward from His (H), and it becomes an important factor to stabilize the His (H) positive side-chain instead of the negative moiety of Asp (D) in the catalytic triad [7,21].
Mechanism of Action of CEs
Esterases catalyze different types of reactions. The primary reaction catalyzed by these enzymes is hydrolysis, but depending upon the reaction conditions, CEs also catalyze esterification, alcoholysis, and transesterification reactions.
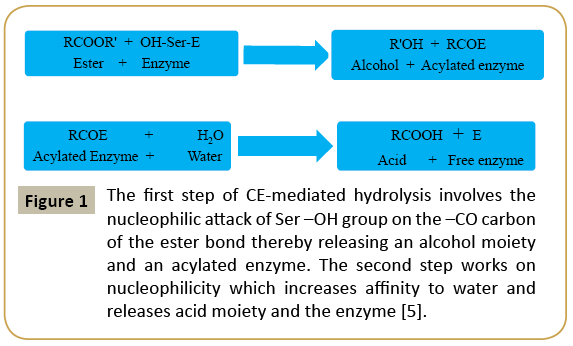
The ability of these enzymes to hydrolyze ester bond to acid and alcohol metabolite is a two-steps process (Figure 1). The first step is nucleophilic attack of catalytic serine –OH on the carbonyl carbon of ester bond. This step releases an alcohol metabolite and an acylated enzyme due to covalent linkages formed between acid moiety of substrate and Ser (S) residue at the catalytic site. The step is stabilized through hydrogen bonding to His (H) which in turn is stabilized by carboxylic group of acidic member of catalytic triad. In the second step, the His (H) residue of the catalytic triad shows affinity toward water molecules and it also helps the enzyme to return to its active state with the release of an acid moiety.
Figure 1: The first step of CE-mediated hydrolysis involves the nucleophilic attack of Ser –OH group on the –CO carbon of the ester bond thereby releasing an alcohol moiety and an acylated enzyme. The second step works on nucleophilicity which increases affinity to water and releases acid moiety and the enzyme [5].
Enzymatic Properties of Carboxylesterases
Among all the enzymes, CEs are the only enzymes which can bear the harsh conditions of industrial biotransformation like high temperature, broad pH range and exposure to denaturing organic solvents. The CEs sourced from natural solvents or microbes are ecofriendly and those of thermophilic microbial origin have natural advantage to perform biocatalysis at temperature above ambient, too.
Hydrolysis of Water Soluble Ester Substrate
The CEs often show inherent specificity towards the substrates of short to moderate chain lengths (C ≤ 12) whereas substrates of longer chain length are poor substrates for these enzymes. Tributyrin (C15H26O6) is a standard substrate to assay the CE activity. The substrate specificity of CEs shows p-nitrophenybutyrate being cleaved by carboxylesterases. Their affinity for short and medium chain length esters is relatively much more than the long carbon chain length esters. This is a criterion employed for their demarcation from lipases.
CEs do not act on the substrates that form micelles
CEs do not appear to act at interface created by hydrophobic lipid substrate(s) and hydrophilic aqueous medium called interfacial activation. However, some of the scientists are of the opinion that the phenomenon of interfacial activation is not a sole distinguishing character between lipases and CEs; e.g., interfacial activation is absent in lipase Lip A from Bacillus subtilis though lid is absent around the active site whereas it is present in lipase Lip A from Pseudomonas aeruginosa [22]. Further the CEs obey Michaelis-Menten equation i.e., these enzymes require minimum substrate concentration for a reaction to occur.
Biochemical Properties of Bacterial Carboxylesterases
Molecular mass of CEs
The most of CEs exist as monomer and oligomers of same subunits with molecular weight broadly ranging from 25 kDa to 85 kDa.
Temperature and pH optima
The CEs show activity in the buffer system with pH raging from alkaline to acidic (pH 3-13) but appear to show maximum activity at pH 7 or 8 i.e., either at neutral or alkaline pH.
Thermophilic, mesophilic and psychrophilic CEs
Bacterial CEs are present at all temperature conditions from moderate to extreme. This enzyme works in the temperature range of 35-65oC but its optimum temperature intriguingly lies at both low as well as high temperature. Highly thermostable CE from hyperthermophilic Pyrobacculum calidifontis VA-1 has a temperature optimum at 90oC while an esterase from Pyrococcus furosus has half-life period of 2 hours at 110oC [4]. Most of hyperthermophilic microbes including archeal sources have been characterized and have completely sequenced genomes. However, very few CEs have been sequenced and characterized from hyperthermophiles sources. The first 3D structure CE that was successfully characterized was from an acidothermophilic bacterium Sulfolobus acidocaldarius [23,24]. CEs also show their presence in halophilic environment [25]. An esterase EstSL3 from Alkalibacterium sp. was the first esterase reported from a coldadapted salted environment of a soda-lake which was capable of functioning both at high pH and high salt concenteration. Such enzymes show adaptation to highly cold environment [13].
Enzyme assays and substrate specificity
The activity of CEs can be measured in terms of a Unit (U) which refers to the amount of enzyme that releases 1 μmol of para-nitrophenol from a para-nitrophenylester (often paranitrophenylesters of C-chain length ≤ 12) per min under specified conditions of temperature and pH [26-28]. Lowering of enzyme activity with the increase in fatty acid chain length is the characteristic of true esterases.
Effect of metal ions on the activity of CEs
To maintain enzyme stability and active enzyme protein structure, divalent metal ions often play an important role. Among divalent cations of Ca, Mg, Mn, Fe, Co, Cu, Cd and Hg, most of these have inhibitory effect on the activity of the esterases, whereas some of these might have stimulatory effect, too. In case of some esterases, the Ca2+, Cd2+ and Mg2+ ions result in better binding of enzyme on to the substrate molecules and also the neutarlisation of fatty acid released from the substrate [29]. In oil-degrading bacterium Acinetobacter lwoffii 16C-1, the addition of divalent cations decreased the activity to greater than 50% resulting in the binding of free fatty acids onto the oil surface thereby increasing their surface area [30]. Some esterases such as PytH pyrethroiddegrading CE from Sphingobium sp. JZ-1 showed little role of metal ions in enzyme activity due to its inactivity towards EDTA, a chelating agent [31].
Carboxylesterase inhibitors
The structure and catalytic properties of carboxylesterases are understood by the use of a variety of inhibitors. CE-inhibitors significantly affected the discovery of anti-esterase drugs (Table 1). The activity of these enzymes is greatly affected by various inhibitors. PMSF, a serine protease inhibitor can covalently link to serine, while diisopropylfluorophosphate (DIFP) and diethylpyrocarbonate (DEPC) are commonly used inhibitor, which inhibit esterase activity thus inferring that Ser (S) as well as His (H) residues are indiscernible for the activity of these enzymes. Change in CEs activity alters drug metabolism and pharmacokinetics and also results in the development of small molecular inhibitors with the purpose of altering drug-induced toxicity. CE inhibitors significantly affected the discovery of drugs [32].
| Substrates | CE producing microorganisms |
|---|---|
| 2-Phenyl acetate (C10H12O2) | Rhodobacteriasphaeoides |
| 4-Nitrophenylacetate (C8H7NO4) | Bacillus subtilis, Geobacillusstereothermophillus, Pseudmonasfluorescens, Arthrobacterglobiformis, Archaeoglobusfulgidus, Alicyclobacillus acidocaldarius, Lactobacillus casae, Klebsiella sp., Sulfolobussolfataricus, Serratia sp. SES-01, SulfolobustokodaiiZD112, Geobacillusstereothermophillus, Streptomyces equiiandThalasobacillussp. |
| 2-Napthyl acetate (C12H10O2) | Pseudomonas fluorescens, Rhodobactersphaeroides, Sulfolobussolfataricus, Thalasobacillussp. Alicyclobacillus tengchogenesisandSufolobussolfataricusMT4 |
| 2-Napthyl butyrate (C14H14O2) | E. coli |
| 2-Napthyl propionate (C13H12O2) | Rhodobactersphaeroides |
| Ethyl chrysanthemate (C12H20O2) | Arthrobacterglobiformis |
| Isopropyidineglycerolbutanoate (C6H12O3) | Bacillus subtilis |
| Ketoprofen ethylester (C6H13N3O2) | Archaeoglobusfulgidus, Pseudomonas fluorescens |
| Naproxen methyl ester (C15H16O3) | Bacillus subtilis, SulfolobussolfatatricusandSulfolobussofataricus P1 |
Table 1: Bacterial carboxylesterases and their substrate specificities.
Effects of detergents and organic solvents on CEs
Esterases due to their inherent stability in organic solvents are of immense use in industrial biotechnology-based application. The ability of these enzymes to reverse the reaction from hydrolysis to synthesis by the removal of water makes them beneficial in various commercial esterification reactions. An esterase from Lactobacillus plantarum, a lactic acid bacterium which is involved in fermented food products shows resistance to organic solvents and metal cations [33]. The detergents being surfaceactive molecules may enhance the spreading/ infusibility of the substrate molecules leading to improved activity of CEs [34].
Classification of CEs
Esterases may be classified based on their substrate specificity (Table 1) as well as their catalytic inhibition pattern (Tables 2 and 3). This classification scheme although did not exactly classify CEs yet it helps to determine biochemical properties of the esterases. In the case of Sulfolobus solfataricus, some inhibitors like PMSF reduces the activity of the enzyme which means that the serine residue(s) at the active site of the enzyme is/ are involved while the inhibition of CE-activity by mercuric chloride indicates the presence of sulfhydryl (-SH) group, too [35].
| S No | Criteria of classification | Types of esterases | References |
|---|---|---|---|
| 1. | Use of inhibition pattern suchas organophosphates paraoxon diisopropylfluorophosphate, fenitrooxon, Carbamate esterinesulphate | a) Acetylesterases not affected by inhibitors and prefer aliphatic substrates b) Arylesterases prefer aromatic substrates and inhibited by sulphydryl reagents. c) Carboxylesterases inhibited by organophosphates (OP) prefer aliphatic substrates d) Cholinesterase simultaneously inhibited by OP and eserinesulphate. |
[5] |
| 2. | International Union of Biochemistry and Molecular Biology (IUBMB) classification (1978) | Aliesterases (Est-B) prefer aliphatic aromatic esterases. Assigned Enzyme Commission number 3.1.1.1 to these enzymes. | [36] |
| 3. | Phylogenetic criteria | Based on sequence similarity of carboxylesterase encoding nucleotide sequences classify them into five families CES 1, 2,3,4,5. Structure and functions. In bacteria, CEs are divided into 8 families which were later expanded to 15 families (Table 2). |
[7,37,38] |
| 4. | Based on primary structure comparison | Lipoprotein lipase (L-family), Hormone sensitive lipase (H-family) Cholinesterase (C-family) |
[1] |
Table 2: Classification given by different researchers.
| Family code | Prominent features | References |
|---|---|---|
| Family I | Lipases with six subfamilies | [7] |
| Family II | GDSL motif with active Ser (S) mostly contain triad conserved at C terminal sequence | |
| Family III | Family III contain majority of lipids only few are esterases | |
| Family IV (HSL) | Pentapeptide GDSAG motif with nucleophilic Ser (S) and HGGG W/F/Y motif | |
| Family V | GLSMG consensus sequence | |
| Family VI | GFSQG, GFSNG conserved sequence | |
| Family VII | GESAG sequence | |
| Family VIII | Ser-X-X-Lys motif in N-terminal part of enzyme | |
| Family IX | GYSLG motif | [39] |
| Family X | GHSLG pentapeptide motif | [23] |
| FamilyXI &XII | Majority of lipases are in both the families | ------ |
| Family XIII | GLSLG pentapeptide motif | [40] |
| Family XIV | CHSMG pentapeptide motif with Cys (C ) residue in place of Gly (G) | [41] |
| Family XV | GLSTG pentapeptide motif instead of common GXSXG motif | [42] |
Table 3: Arpigny and Jaegar classification scheme for microbial CEs.
Based upon the specificity of the enzyme for its substrate (chain length), GDSL motif, triad nature of catalytic site, nucelophilic serine and consensus sequences, the esterases have been classified into I to XV families (Table 3).
Family I
The family I contains true lipases and this family is further subdivided into six subfamilies.
Family II
The CEs in this family show GDSL motif instead of classical GXSXG present in most of esterases. These enzymes have been reported from Streptomyces scabies, Pseudomonas aeruginosa, Salmonella typhimurium, Ferbidobacterium nodusum Rt 17 B-1 and an esterase EstSL3 from Alkalibacterium sp. [43]. The esterase from Streptomyces scabies lacked Asp (D) and contained a diad of Ser (S) and His (H), instead of a typical catalytic triad is a member of this family. The backbone carbonyl of the residue located three residues upward from His (H) become a stabilizing factor to stabilize His (H) positive side chain instead of negative chain of Asp (D) in catalytic triad [7,21]. GDSL family is further subdivided based on conservation of S, G, H and D residues into four blocks I, II III and IV. The enzymes in this family possess different functional properties and broad substrate specificity due to flexible active catalytic site that undergoes induced-fit conformational change [44,45].
Family III
These CEs are characterized by a triad Ser (S), Asp (D) and His (H) which have functional resemblance with subtilisin and trypsin. The family consists of majority of lipid hydrolyzing CEss. MtEst45 from Microbulbifer thermotolerans DAU221 from this family is categorized as an esterase although its substrate kinetics shows maximum activity with carbon chain length of 4 and poor activity with C chain length of 2 and 6 [29].
Family IV
These esterases are called hormone-sensitive lipases because of their primary amino acid sequence shows similarity with that of hormone sensitive lipases. The active site Ser (S) is the part of conserved sequence pentapeptide motif GDSAG at position Ser-157, His-254 and Asp-284 and an oxyanion hole having sequence motif HGGG F/W/Y, which are involved in hydrogen bonding interactions and also stabilize the transition state during the hydrolysis. There is a distinct sequence similarity between esterases from mesophilic, thermophilic and psychrophilic CEs. Some of the esterases at thermophilic and extreme ends from this family are Archaeoglobus fulgidus [46], Sulfolobus solfataricus P1 [47], Pyrobacculum calidifontis [4], Bacillus acidocaldarius [48], Est ATII; as well as halotolerant forms whereas its mesophilic counterparts are Lactobacillus plantarum [49], Oenococcus oeni [50], Acinetobacter lwoffii 16 C-1 [30], and some members are also isolated from metagenomic sources [14,51,52]. The metalloresistant Est ATII is a thermophilic as well as halotolerant esterase reported from extremely harsh conditions [53]. Est J was the first alkaliphilic and moderately thermophilic esterase and first thermoalklostable 499EST esterase has been reported from gram negative bacterium Acidicaldus sp. strain USBA-GBX-499 [54]. Proteins present in this family are monomers and dimers as in Pest E from Pyrobacculum calidifontis VA-1 and octamers also as is the case of esterase AFEst from Archaeoglobus fulgidus. The active site as well as substrate binding site is complete within monomer. All HSL proteins in their structural form show same processive orientation of substrate in the active site [55]. An esterase from Pyrobacculum calidifontis VA-1 is the most stable thermophilic esterase among all the esterases reported from this family. This esterase can sustain its activity for 2 h at 100°C.
Family V
The enzymes in this family originate from mesophilic bacteria such as Acetobacter pasteureannus, Pseudomonas oleororans and Haemophilus influenza as well as from cold-adapted Sulfolobus acidocaldarius. Only a few esterases have been characterized from this family.
Family VI
These esterases are smallest known esterases having molecular weight of 23-26 kDa. The crystal structure of an esterase from Pseudomonas fluorescence has been successfully determined [20]. Est A6 from a Pseudomonas sp. CR-611 [56] belonged to this family. A pentapeptide motif GXSXG with catalytic triad of Ser-Asp- His hydrolysing small substrates is present in these esterases. CEs in this family are present as active homodimeric proteins. Highly alkal tolerant, halotolerant and enatioselective Est PE8 from marine Pelagebacterium halotolerans B2T is a potential source of a precursor (R)-3-MFG used for the synthesis of anti-depressant paroxetine hydrochloride. Among all the esterases, this family is known for its smallest (23-26 kDa) molecular mass [57]. Another esterase from Spirulina platensis [58] and pyrethroid hydrolysing esterase from Ochrobactrum anthropi YZ-1 have also been reported from this family [59].
Family VII
Esterases from this family show similarity to acetylcholiesterase from the eukaryotes and intestine liver carboxylesterase. The sequences show the conserved pentapeptide motif GESAG. The Bacillus subtilis esterase was found to hydrolyze para-nitrobenzyl esters more effectively. The esterase sequences from this family encode proteins of molecular mass 60-65 kDa and are largest known esterases. These enzymes have ability to sterically hindered esters of tertiary alcohol and polyurethanes which are widely used in the industry. The enzymes from this family are characterized from compost metagenomic library [60].
Family VIII
The members belonging to this family show high sequence identity to peptidases and β-lactamases as is true carboxylesterase from Brevibacterium linens IFO12171 [61]. The primary sequence of these enzymes show conserved sequence motif S-X-X-K at the N-terminal whereas C-terminal of these enzymes also have a conserved sequence motif G-X-S-X-G. The S-X-X-K motif is conserved in peptidases and β-lactamases also [62-65]. Jeon et al. reported an esterase EstU1 from soil metagenome which has the same active site residue(s) to be involved in ester bond (PNP ester) as well as amide bond hydrolysis (β-lactamases). This unique catalytic action of the enzyme was supported by site directed mutagenesis [27]. The enzymes from this family are reported from different environments including marine sponge, cold adapted and metagenomes obtained from compost and leachate environment [29,66]. Most of carboxylesterases included in this family are from metagenomes. Mokoena et al. referred the esterases from this family as poorly characterized esterases [67]. Est C represents the first member of Family-VIII esterases with the unique β-lactam hydrolytic activity [68].
Family IX
This family came into existence with a characterized thermostable esterase HydS14 obtained from actinobacteria Actinomadura sp. strain S14. The esterase was cloned and expressed for the first time in Pichia pastoris instead of commonly used E. coli as an expression vector. The encoded proteins have a common pentapeptide motif GYSLG [69].
Family X
An esterase EST D from hyperthermophilic Thermotoga maritime is the first characterized member of this family. Proteins are present in monomeric form. It is a second most stable bacterial esterase with a half-life of one hour at 100oC [23].
Family XI and XII
Both these families consist of majority of lipases.
Family XIII
All the members in this family are derived from Bacillus sp. The other members of this family include EstOF4 from Bacillus pseudofirmus. The enzymes are moderately thermostable and are adapted to extreme alkaline environment (pH 11). The Proteins in native form are dimers and are hydrophobic [40].
Family XIV
This family came into existence with the thermostable esterase EstA3 from Thermoanaerobacter tengcogenesis MB4. The enzyme sequence contains a pentapeptide motif CHSMG with Cys (C) residue at the first position [41].
Family XV
The enzymes from this family contain ‘GLSTG’ a distinct pentapeptide motif which is quite different from the common GXSXG motif found in most of the esterases. The esterase(s) from Geobacillus thermodenitrificans strain CMB-A2 and Geobacillus thermoleovoras CCR11 in this family show resistance to temperature, pH and organic solvents and is therefore found to be very amenable in the processes like oleochemical biotransformations, pharmaceuticals, cosmetics, organic synthesis and biodiesel production [42,70,71].
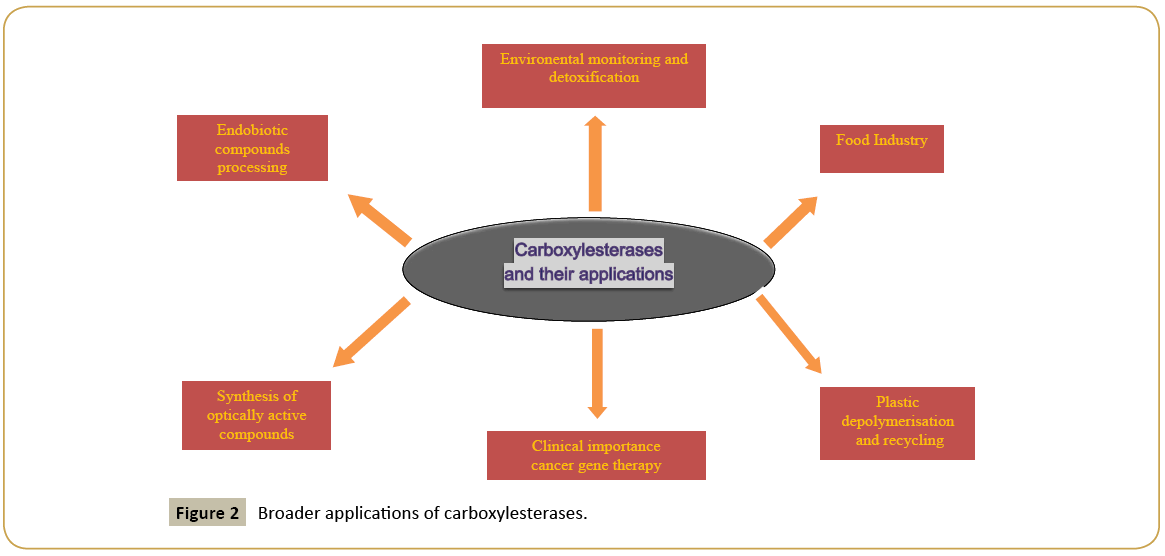
Applications of CEs
The extensive uses of these enzymes result in the search of novel sources of CEs with functional values suited well for their industrial applications. These enzymes are well known for their role in green chemistry.
Detoxification reactions and environmental monitoring
Insecticides can be degraded by chemical as well as microbial interventions. Artificially created pesticides are often ester compounds. Microbial breakdown and environmental reactions like hydrolysis, photolysis and oxidation with minerals are some of the reactions that convert pesticides into nontoxic or less toxic compounds. Microbial breakdown is least harmful, safe, ecofriendly and economic method exploiting the application of CES (Figure 2). Pyrethroids and malathion are organophosphorus insectisides having harmful effects on human beings and other higher organisms [72-74]. Enzymes from Alicyclobacillus tengchogenesis degrade malathion quite efficiently. Malathion also acts as a sole carbon source for the bacterial species grown in minimal salt medium such as Bacillus licheniformis helps in the bioremediation of soil that is polluted with malathion [75,76]. The degradation of malathion by Brevibacillus sp. and Bacillus cereus has also been reported [77]. Toxicity caused due to OPs results in the inhibition of acetylcholiesterases (a neurotransmitter) resulting in the disruption of the nervous system in the rats and other pests due to formation of neurotransmitter acetylcholine. CEs bind stochiometrically to these chemicals. Cleavage of pyrethroides by CEs is also one of the detoxification pathways in mammals and insects which possess isomer selectivity. Carboxylesterase gene(s) from Bacillus cereus SM3, Aspergilus niger ZD11, Nephotettix cincticeps and mouse liver microsomeshydrolyzing carboxylester linkages in pyrethroids have been successfully purified. The CE genes from liver microsomes and those from Klebsiella sp. strain ZD112 were cloned and functionally expressed [72].
Endobiotic compounds processing
CEs play important roles in endobiotic processing of hydrophobic compounds e.g., cholesterol, which is an important constituent involved in maintaining the structural integrity of plasma membrane. Its excess causes a harmful effect though its normal quantity plays a vital role in cholesterol homeostasis in the animals. CEs transform fatty acid at 3rd position of the cholesterol to form cholesteryl ester.
Biocatalysis
Besides detoxification reactions, CEs are involved in bio catalysis, a chemical reaction in which one or more enzymes catalyze the chemical reaction, which find potential applications in synthetic chemistry.
Synthesis of optically active compounds
CEs are also involved in bio catalysis especially in organic compounds synthesis. Their use in ‘White Biotechnology’ for the synthesis of industrially important chiral compounds is increasingly important in pharmacology. The drugs that commonly enter in the market consist of one of its either of two isomers. Due to their high region-stereo specificity, CEs are used in the synthesis of optically pure compounds. One of such compounds i.e., carboxylesterases NP (Naproxen) from Bacillus subtilis Thai I-8 was characterized as a very effective enantioselective biocatalyst for NSAID. Naproxen is non-steroidal, pain killing drug with only one of its optical isomer that is commonly used [77]. The CEs show modest selectivity towards chiral alcohol as compared to carboxylic acids. It is produced as an intracellular protein with a molecular weight of 32 kDa, with optimum pH of 8.5-10.5 and optimum temperature between 35 to 55°C. Besides carboxylesterase NP, 2-arylpropanoic acid is produced with high enantioselectivity [1°]. An esterase from Arthrobacter globiformis is used in the chemical synthesis of (+)-trans-(1R, 3R) - chrysanthemic acid, which is an important precursor of pyrethrin insecticides [78].
Food industry
As food additives, enzymes act as important means to enhance the flavor, texture and taste of the foods. Esterases are commonly used in making cheese and other milk derivatives and their important role is considered in improving aroma, taste and texture of many dairy products. Esters are also important for the aroma of fermented beverages like wine with ethyl acetate as the most common ester in wine due to its ready formation from ethanol and acetic acid. High reactivity of a primary alcohol and its low concentration gives specific fruity character to the wine [50]. The first CE cloned and characterized from wine was associated with bacterium Oenococcus oeni. Esters from Pseudomonas fragii and Ferbidobacterium nodusum Rt 17 B-1 are responsible for fragrance and fruit-like flavours. Free fatty acids in cheese are important for flavor. CEs play important role in apple flavor and their biosynthesis increases with the fruit ripening hormone ethylene. During early stages of fruit development these enzymes are expressed at low levels. At maturity their expression level increases [79]. These enzymes also play role in the chemical modification of many drugs and prodrugs like antiplatelet drug aspirin, clopidogrel and drugs like delapril, imadapril, and temocapril which are acetylcholiesterase inhibitor and an antitumor drug irinotican [80].
Cancer gene therapy
CEs have been implicated in the activation of a number of anticancer agents like CPT-11, irinotican [7-ethyl-10-[4-(1- piperidino)-1-piperidino] and carbonyoxycamptothecin, which are active against a broad range of cancers including colorectal and cervical cancer. Anticancer effect of CPT-11 is dependent on its conversion to ethyl camptothecin, which is catalysed by one or more CEs particularly CE-2. These enzymes play role to activate prodrugs in vivo and generate effective anticancer agents in highly selected target site at the surface of tumor cells or inside them CEs are targeted to tumor sites with hybrid monoclonal antibodies or cDNA encoding a CE is targeted to tumor cells by viral vector. This therapeutic strategy releases anticancer drug ethyl camptothecin in the vicinity of tumor cells [79], thus killing them effectively.
Plastic depolymerization and recycling
Microbial CEs are effective biocatalysts and can hydrolyze various polyesters effectively and play an important role in plastic depolymerization like polylactic acid (PLA). Recycling of PLA is required which produces CO2 [81,82]. Tchigvintsev et al. reported that metagenomic esterases have ability to hydrolyze many polyester substrates including PLA [15].
Conclusion
Ecofriendly nature and various other applications of CEs such as their role in drug metabolism, food industry and clinical use prove them to be beneficial. Because of their use in xenobiotic compounds processing (especially OPs) and insecticide-resistance in microorganisms, these enzymes are emerging as the basis of various bioremediation methods to lessen the effect of xenobiotic environmental pollutants. Further studies on these enzymes will not only help to explore new strategies of pollution control methods but will also provide diverse industrial applications for human welfare.
Acknowledgements
The financial assistance in the form of Research Fellowship to AS (DST/INSPIRE Fellowship/2013/1036) and SS (Research Fellowship funded by DBT, New Delhi are thankfully acknowledged.
Conflict of Interest
None of the authors have conflict of interest, financial or otherwise among themselves or with the parent institute.
References
- Chahinian H, Sarda L (2009) Distinction between esterases and lipases: comparative biochemical properties of sequence related carboxylesterases. Protein Pept Lett 16: 1149-1161.
- Sharma S, Kanwar SS (2014) Organic solvent tolerant lipases and applications. Scientific World J p: 625258.
- Fojan P, Jonson PH, MariaTNP, Steffen BP (2000) what distinguish an esterase from a lipase: A novel structural approach. Biochimie 82: 1033-1041.
- Hotta Y, Ezaki S, Atomi H, Imanaka T (2002) Extremely stable and versatile carboxylesterase from a hyperthermophilic Archaeon. Appl Environ Microbiol 68: 3925-3931.
- Montella IR, Schama R, Valle D (2012) the classification of esterases: An important gene family involved in insecticide resistance- A Review. Mem Inst Oswaldo Cruz 107: 437-449.
- Xie G, Liu M, Zhu H, Lei B (2008) Esterase SeE of Streptococcus equissp.equi is a novel nonspecific carboxyl ester hydrolase. FEMS 289: 181-186.
- Arpigny JL, Jaegar KE (1999) Bacterial Lipolytic Enzymes: Classification and properties.Biochem 1:177-83.
- Lopes DB, Fraga LP, Fleuri LF, Macedo GA (2011) Lipase and esterase-to what extent can this classification be applied accurately. CiencTechnol Aliment Campinas 31:608-613.
- Aldridge WN (1953) Serum esterases. I. Two types of esterase (A and B) hydrolysing p-nitrophenyl acetate, propionate and butyrate and a method for their determination. Biochem 53: 110-117.
- Bornscheuer UT (2002) Microbial carboxyl esterases: classification, properties and applications in biocatalysis. FEMS Microbiol Rev 26:73-81.
- Sayali K, Sadichha P, Surekha S (2013) Microbial Esterases: An Overview.CurrMicrobiol App Sci 2:135-146.
- Hosokawa M(2008) Structure and catalytic properties of carboxylesterase isozymes involved in metabolic activation of prodrugs. Molecules 13:412-431.
- Wang G, Wang Q, Lin X, Ng TB, Yan R, et al. (2016) A novel cold-adapted and highly salt-tolerant esterase from Alkalibacterium sp. SL3 from sediment of a soda lake. Sci Rep 6.
- Rashamuse K, Ronneberg T, Hennessy F, Visser D, Van Heerden E, et al. (2008) Discovery of a novel carboxylesterase through functional screening of a pre-enriched environmental library. ApplMicrobiol 106: 1532-1539.
- Tchigvintsev A, Tram H, Popovic A, Kovakik F, Brown G, et al. (2015) the environment shapes microbial enzymes: five cold active and salt resistant carboxylesterases from marine metagenomes. ApplMicrobiolBiotechnol 99: 2165-2178.
- Kang CH, Oh KH, Lee MH, Oh TK, Kim BH (2011) A novel family VII esterase with industrial potential from compost metagenomic library.Microb Cell Fact 10:41.
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, et al. (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobusfulgidus Nature 390: 364-370.
- Sanghani SP, Sanghani PC, Schiel MA, Bosron WF (2009) Human carboxylesterases: an update on CES1, CES2 and CES3. Protein Pept Lett 16:1207-1214.
- Holmes RS, Cox LA,Vandeberg JL (2009) A new class of mammalian carboxylesterase CES6 Comp BiochemPhysiol Part D. Genomics Proteomics 4: 209-217.
- Kim KK, Song HK, Shin DH, Hwang KY, Choe S, et al. (1997) Crystal structure of carboxylesterase from Pseudomonas fluorescens, an α/β-hydrolase with broad substrate specificity. Structure 5:1571-1584.
- Wei Y, Schottel JL, Derewenda U, Swenson L, Patkar S, et al. (1995) A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat Struct Biol 2:218-223.
- Hausmann S, Jaegar KE (2010) Lipolytic enzymes from bacteria. In: Handbook of hydrocarbon and lipid microbiology. Springer-Verlag Berlin, Heidelberg, Germany.
- Levisson M, Oost JVD, Kengen SWM (2007) Characterization and structural modeling of a new type of thermostable esterase from Thermotogamaritime. FEBS 11:2832-2842.
- Kim S, Lee SB (2004) Thermostable esterase from a thermoacidophilic archaeon: purification and characterization for enzymatic resolution of a chiral compound. BiosciBiotechnolBiochem 68: 2289-2298.
- Jiang H, Zhang S, Gao H, Hu N (2016) Characterization of a cold-active esterase from Serratiasp. And improvement of thermostability by directed evolution. BMC Biotechnol 16:7.
- Rhee JK, Kim DU, Ahn DG, Yun JH (2006) Analysis of the thermostability determinants of hyperthermophilic esterase EstE1 based on its predicted three dimensional structure. Appl Environ Microbiol 72:3021-3025.
- Jeon JH, Kim SJ, Lee HS, Cha SS, Lee JH, et al. (2011) Novel metagenome-derived carboxylesterase that hydrolyzes β-lactam antibiotics. Appl Environ Microbiol 77:7830-7836.
- Sharma T, Sharma A, Kanwar SS (2016) Purification and characterization of an extracellular mass esterase from Bacillus pumilus. J Advance BiotechnolBioengg 4: 9-16.
- Lee YS (2016) Isolation and characterization of a novel cold adapted esterase, MtEst45, from Microbulbiferthermotolerans DAU221. Front Microbiol 7:218.
- Kim HE, Park KR (2002) Purification and characterization of an esterase from Acinetobacter lwoffii 16C-1. CurrMicrobiol 44:401-405.
- Zhu Y, Li J, Cai H, Ni H (2013) Characterization of a new thermostable esterase from metagenomic library. Elsevier 168:589-597.
- Hatfield MJ, Potter PM (2011) Carboxylesterase inhibitors. Expert OpinTher Pat 21:1159-1171.
- Inmaculada NZ, Alvaro SF, Francisco GC (2013) Overexpression, purification, and biochemical characterization of the esterase Est0796 from Lactobacillus plantarum WCFS1. MolBiotechnol 54: 651-660.
- Zhang T, Han WJ, Liu ZP (2009) Gene cloning and characterization of a novel esterase from activated sludge metagenome. Microb Cell Factories pp:1-8.
- Kim, Lee (2004) Thermostable esterase from a thermoacidophilic archaeon: purification and characterization for enzymatic resolution of a chiral compound. BiosciBiotechnolBiochem 68: 2289-2298.
- Walker CH (1993) the classification of esterases which hydrolyse organophosphates: recent developments. ChemBiol Interact 87: 17-24.
- Satoh T, Hosokawa M (1995) Molecular aspects of carboxylesterase isoforms in comparison with other esterases. Toxicol Let 83: 439-445.
- Satoh T, Hosokawa M (2006) Structure, function and regulation of carboxylesterases. ChemBiol Interact 162: 195-211.
- Handrick R, Reinhardt S, Focarete MT, Scandola M, Adamus G, et al. (2001) A new type of thermoalkalophilic hydrolase of Paucimonaslemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J BiolChem 276: 36215-36224.
- Rao L, Xue Y, Zheng Y, Lu JR, Ma Y (2013) A novel alkaliphilic Bacillus esterase belongs to 13th bacterial lipolytic enzyme family. PLoS ONE 8:e60645.
- Rao L, Xue Y, Zhou C, Tao J, Li G, et al. (2011)A thermostable esterase from Thermoanaerobactertengcongensis opening up a new family of bacterial lipolytic enzymes. BiochimBiophys Acta 1814:1695-1702.
- Luna G E, Sanchez-Otero MG, Castro RQ, Toledo REM, Ros RMO (2016) Gene Cloning and Characterization of the Geobacillusthermoleovorans CCR11 carboxylesterase CaesCCR11, a new member of family XV.MolBiotechnol 58: 37-46.
- Wicka M,Wanarska M, Krajewska E, Anna PS,Kur J, et al. (2016) Cloning, expression, and biochemical characterization of a cold active GDSL-esterase of a Pseudomonas sp. S9 isolated from Spitsbergen island soil. Acta Biochim63:117-125.
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43: 534-552.
- Yu EY, Kwon MA, Lee M, Oh JY, Choi JY, et al. (2011) Isolation and characterization of cold active family VIII esterases from an arctic soil metagenome. ApplMicrobiolBiotechnol 90:571-581.
- De Simone G, Menchise V,Manco G,Mandrich L, et al. (2001) The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobusfulgidus.MolBiol 314: 507-518.
- Park YJ,Choi SY, Lee HB (2006) A carboxylesterase from the thermoacidophilic archaeon Sulfolobussolfataricus P1; purification, characterization, and expression.BiochimBiophys Acta 1760: 820-828.
- Manco G, Febbraio F, Adinolfi E, Rossi M (1999) Homology modeling and active-site probing of thermophilic Alicyclobacillus acidocaldarius esterase 2. Protein Sci 8:1789-1796.
- Brod FCA, Vernal J, Bertoldo JB, Terenzi H, Arisi AC (2010) Cloning, expression, purification, and characterization of a novel esterase from Lactobacillus plantarum. MolBiotechnol 44: 242-249.
- Sumby KM, Matthews AH, Grbin PR, Jiranek V (2009) Cloning and characterization of intracellular esterases from the wine- associated Lactic acid bacterium Oenococcusoeni.Appl Environ Microbiol 75: 6729-6735.
- Ko KC, Rim SO, Han Y, Shin BS, Choi JH, et al. (2012) Identification and characterization of novel cold adapted esterase from metagenomic library of mountain soil.IndMicrobiolBiotechnol 39: 681-689.
- Fu C, Hu Y, Xie F,Guo H, Ashforth EJ, et al. (2011) Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. ApplMicrobiolBiotechnol 90:961-970.
- Mohamed YM, Ghazy MA, Sayed A, Ouf A, Dorry HEI, et al. (2013) Isolation and characterization of a heavy metal resistant, thermophilic esterase from a red sea brine pool. Sci Rep 3: 3358.
- Choi EJ, Kwon MA, Na HY, Hahm DH, Song JK (2013) Isolation and characterization of a metagenome-derived thermoalkaliphilic esterase with high stability over high pH range. Extremophile 17: 1013-1021.
- Palm GJ, Alvaro EF, Bogdanovic X, Bartsch S, SczodrokJ, et al. (2011) The crystal structure of an esterase from the hyperthermophilic microorganism Pyrobacculumcalidifontis VA1 explains its enantioselectivity. ApplMicrobiolBiotechnol 91: 1061-1072.
- Prim N, Bofill C, Pastor FIJ, Diaz P (2006) Esterase EstA6 from Pseudomonas sp. CR-611 is a novel member in the utmost conserved cluster of family VI bacterial lipolytic enzymes. Biochimie88: 859-867.
- Wei X, Jiang X, Ye L, Yuan S (2013) Cloning, expression and characterization of a new enatioselective esterase from a marine bacterium Pelagibacteriumhalotolerans B2T. MolCatal B Enzym 97:270-277.
- Salvi S, Trinei M, Lanfaloni L, Pon CL (1994) Cloning and characterization of the gene encoding an esterase from Spirulina platensis.Mol Gen Genet 243: 124-126.
- Zhai Y, Li K, Song J, Shi Y, Yan Y (2012) Molecular cloning, purification and biochemical characterization of a novel pyrethroid-hydrolyzing carboxylesterase gene from OchrobactrumanthropiYZ-1. Hazard Mater 222:206-212.
- Tao W, Lee MH, Wu J, Kim NH, Lee SW (2011) Isolation and characterization of family VII esterases derived from alluvial soil metagenomic library. Microbiol 49:178-185.
- Sakai Y, Ishikawa J, Fukasaka S, Yurimoto H, Mitsui R, et al.(1999) A new carboxylesterase from Brevibacterium linens IFO12171 responsible for conversion of 1,4-Butanediol Diacrylate to 4- Hydroxybutyl Acrylate: Purification, characterization, gene cloning and gene expression in Escherichia coli. BiosciBiotechnolBiochem 63:688-697.
- Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, et al. (2006) Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol 72: 3637-3645.
- Kim YH, Kwon EJ, Kim SK, Jeong YS, Kim J, et al. (2010) Molecular cloning and characterization of a novel family VIII alkaline esterase from a compost metagenomic library. BiochemBiophys Res Commun 393: 45-49.
- Yu EY, Kwon MA, Lee M, Oh JY, Choi JY, et al. (2011) Isolation and characterization of cold active family VIII esterases from an arctic soil metagenome. ApplMicrobiolBiotechnol 90:571-581.
- Cha SS, An YJ (2016) Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins. BiochemBiophys Res Comm 478: 818-824.
- Rashamuse K, Magomani V, Ronneburg T, Brady D (2009) a novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous β-lactamase activity on nitrocefin.ApplMicrobiolBiotechnol 83: 491-500.
- Mokoena N, Mathiba K, Tsekoa T, Steenkamp P, Rashamuse K (2013) Functional characterisation of a metagenome derived family VIII esterase with a deacetylation activity on β-lactam antibiotics. BiochemBiophys Res Commun 437: 342-348.
- Cheng X, Wang X, Qiu T, Yuan M (2014) molecular cloning and characterization of a novel cold-adapted family VIII esterase from a biogas slurry metagenomic library. MicrobiolBiotechnol 24: 1484-1489.
- Sriyapai P, Kawai F, Siripoke S, Chansiri S, Sriyapai T (2015) Cloning, Expression and Characterization of a thermostable esterase HydS14 from Actinomadura sp. Strain S14 in Pichia pastoris. Mol Sci 16:13579-13594.
- Pesaresi A, Devescovi G, Lamba D, Venturi V,Degrassi G (2005) Isolation characterization, and heterologous expression of a carboxylesterase of Pseudomonas aeruginosa PAO1. CurrMicrobiol50: 102.
- Charbonneau DM, Meddeb-Mouelhi F, Beauregard M (2010) a novel thermostable carboxylesterase from Geobacillusthermodenitrificans: evidence for a new carboxylesterase family. Biochem 148: 299-308.
- Wang BZ, Gou P, Hang BJ, Li L, He J, et al. (2009) Cloning of a novel pyrethroid hydrolyzing carboxylesterase gene from Sphingobium sp. Appl Environ Microbiol 75: 5496-5500.
- Goda SK, Elsayed IE, Khodair TA, EIayed W, Mohamed ME (2010) Screening for and isolation and identification of malathion-degrading bacteria: cloning and sequencing a gene that potentially encodes the malathion-degrading enzyme, carboxylesterase in soil bacteria. Biodegradation 21: 903-913.
- Khan S, Zaffar H, Irshad U, Ahmad R, Khan AR, et al. (2016) Biodegradation of Malathion by Bacillus licheniformis strain ML-1. Arch Biol Sci Belgrade 68:51-59.
- Xie Z, Xu B, Ding J, Liu L, Zhang X, et al. (2013) Heterologous expression and characterization of a malathion-hydrolyzing carboxylesterase from a thermophilic bacterium, Alicyclobacillus tengchogensis. Biotechnol Lett 35: 1283-1289.
- Singh B, Kaur J, Singh K (2012) Biodegradation of malathion by Brevibacillus sp. strain KB2 and Bacillus cereus strain PU. World J MicrobiolBiotechnol 28: 1133-1141.
- Littlechild JA (2015) Archaeal enzymes and applications in industrial biocatalysts. Archaea J.
- Nishizawa M, Shimizu M, Ohkawa H, Kanaoka M (1995) Stereoselective production of (+)-transchrysanthemic acid by microbial esterase: cloning, nucleotide sequence, and overexpression of esterase gene from Arthrobacterglobiformis in Escherichia coli. Appl Environ Microbiol 61: 3208-3215.
- Souleyre EJF, Marshall SDG, Oakeshott JG, Russell RJ, Plummer KM, et al. (2011) Biochemical characterization of MdCXE1, a carboxylesterase from apple that is expressed during fruit ripening. Phytochem 72: 564-571.
- Xu Y, Zhang C, He W, Liu D (2016) Regulations of xenobiotics and endobiotics on carboxylesterases: a comprehensive review. Eur J Drug MetabPharmokinet 41:321-330.
- Hsieh YT, Chen KC, Cheng CM, Cheng TL, Tao MH, et al. (2015) Impediment to enhancement of CPT-11 anticancer activity by E.coli directed beta-glucuronidase therapy. PloS One 10:e0118028.
- Hajighasemi M, Boguslaw PC, Tchigvintsev A, Brown G, Flick R, et al.(2016) Biochemical and structural insights into enzymatic depolymerization of polylactic acid and other polyesters by microbial carboxylesterases. Biomac 17:2027-2039.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences