ISSN : 0976-8505
Der Chemica Sinica
Synthesis, Characterization and In-Silico ADMET Screening of Mono- and Dicarbomethoxylated 6,6'-Methylenebis(2-cyclohexyl-4-methylphenol) and Their Hydrazides and Hydrazones
Akbar Ali1, Mohamed EI Badawy2, Raza Shah1, Wajid Rehman1, Yeldez El kilany3, El Sayed H El Ashry1,3* and Nawaz Tahir4
1International Centre for Chemical and Biological Sciences, H.E.J Research Institute of Chemistry, University of Karachi, Pakistan
2Department of Pesticide Chemistry and Technology, Faculty of Agriculture, 21545-El-Shatby, Alexandria University, Egypt
3Chemistry Department, Faculty of Science, Alexandria University, Alexandria, Egypt
4Department of Physics, University of Sargodha, Sargodha, Pakistan
- Corresponding Author:
- El Sayed H El Ashry
International Centre for Chemical and Biological Sciences
H.E.J Research Institute of Chemistry
University of Karachi, Pakistan
Abstract
Mono-carboxymethylation and the dicarboxymethylation of 6,6'-methylenebis(2-cyclohexyl-4-methylphenol) has been accomplished to give methyl-2- (2-cyclohexyl-6- (3-cyclohexyl-2-hydroxy-5-methyl benzyl)-4-methyl phenoxy) acetate (2) and dimethyl 2,2`-(6,6`-methylenebis(2-cyclohexyl-4-methyl-6,1 phenylene) bis(oxy)diacetate (5), respectively. Their structures have been determined using spectroscopic methods. Hydrazinolysis of 2 and 5 gave the corresponding hydrazides, whose reactions with a number of aldehydes gave the Schiff bases. These hydrazones are of considerable interest as precursors for the synthesis of heterocyclic compounds of hybrid nature in addition to their potential biological activity. Pharmacophore elucidation of the compounds was performed based on ligand alignment. In adition, in silico ADMET analysis was done to predict the ADMET properties of the test compounds.
Keywords
Methylenebis-phenoles; Selective alkylation; Acylhydrazones; Pharmacophore; ADMET screening; Docking
Introduction
Heterocycles has a key role in pharmaceuticals, natural products, analytical reagents, agriculture products, and dyes. Much more attention has been paid to develop new methodologies to get these important heterocyclic compounds. To choose a suitable starting material has a pivotal role in the synthesis of novel heterocycles, substituted with unique functional groups. 2,2'-Methylenebis(4-methylphenol) has an important role in many synthetic processes not only because of its electrical and damping behaviours but also its role as precursor to get important heterocycles such as azacalixarenes [1,2], macro molecules like Calix arenes [3], bulky titanium bis(phenolate) complexes which act as initiators for living anionic polymerization of õ-caprolactone [4], stable oligomethylenephenolcyclophosphites containing 1-3 phosphorus atoms which allow design complex coordination systems holding promise for metal complex catalysis [5], dinuclear aluminum complexes supported by amino or imino-phenolate ligands [6], anion receptors based on acyclic phenol-formaldehyde oligomers bearing thiourea groups and has a role in complexation [7]. From the medicinal point of view, 2,2'-methylenebis(4-methylphenol) has a strong role such as inhibition of the sarco/endoplasmic reticulum calcium ATPase [8] and scolicidal effect against Echinococcus granulosus in vitro [9]. Undisputedly acyl hydrazones have also numerous applications in terms of their potential biological activities such as anticonvulsant [10], antidepressant [11], anti-inflammatory [12,13], antimalarial [14,15], antimycobacterial [16,17], anticancer [18,19], antimicrobial [20-23], trypanocidal [24], anti-HIV [25], antiviral and influenza virus agonists [26]. Due to the large number of applications of the products and their precursor both on synthetic as well as medicinal value, we have selected 2,2'-methylenebis(4-methylphenol) to synthesize a novel series of hydrazones for the exploration of interesting bioactive compounds based on diverse structural features, easy synthetic roots and the desired functionalities in the molecules.
The QSAR/QSPR community has, for a good number of years, developed models for the prediction of physicochemical properties of interest in ADMET (absorption, distribution, metabolism, excretion, and toxicity). These include: the partition coefficient, aqueous solubility [27], absorption and permeability [28], BBB penetration [28], plasma protein binding [28], metabolism [29], hERG inhibition [30], excretion [28], P-glycoprotein (P-gp) efflux, physiologicallybased pharmacokinetic (PBPK) modelling and toxicity [31,32]. In addition, of course, pharmacophore and homology modelling have also proceeded, to allow improved prediction of metabolism and toxicity [33,34]. Today, the tests that make up ADMET evaluation are low throughput, and are apparently not informative or accurate enough to predict a drug's probability of success, given the high failure rate of compounds at all stages of development [35]. Drug discovery companies are therefore seeking to reorganize the ADMET process, advancing the chain of early discovery. The objective is to predict, early in the process, perhaps even before the compounds are synthesized, which compounds pass the test for a good drug. Over the past few years, many software have been developed for the properties and toxicity of ADME-based organisms. Software is now available for BBB penetration, human intestinal absorption, oral bioavailability, jejunum permeability, Caco-2 and Madin–Darby canine kidney cell permeability, serum protein binding, apparent volume of distribution, hERG potassium channel blockage, P-gp efflux, human hepatotoxicity, carcinogenicity, mutagenicity, skin sensitisation, develop mental toxicity, metabolism, CYP inducers, substrates and inhibitors, and PBPK [36].
The present work was done on the selective and full carbethoxymethylation of 6,6`-methylenebis(2-cyclohexyl-4- methylphenol in order to use them as precursors for hydrazides and then heterocycles. The single crystal analysis of both the aromatic ester shows that the crystals are stabilized by Van der Waals forces. Since the in vivo and in vitro evaluations are costly and laborious, in silico techniques have been widely used to estimate these properties. In this research study, we would briefly describe the advances of in silico ADMET prediction, with emphasis on structure pattern recognition that was developed recently. The in silico ADMET, pharmacokinetics and drug-likeness prediction were discussed in details and the computational data could be used for prediction in drug discovery and hazard risk assessment.
Materials and Methods
General methods and materials
Melting points were determined with a Mel-Temp apparatus (SMP10) in open capillaries and are uncorrected. TLC was performed on E. Merck Silica Gel 60 F254 with detection by UV light absorption. FT-IR was done on Perkin Elmer Spectrometer.1H NMR spectra were recorded on BrukerAvance AV NMR spectrometer at 300 or 400 MHz, whereas the 13C NMR was recorded on the same instrument at 75 or 100 MHz, respectively, with TMS as internal standard. Mass spectra were recorded on a Finnigan (MAT312) and Jeol (JMS.600H) instrument; HRMS were recorded with Thermo Finnegan (MAT 95XP). Solvents used were purified by simple distillation.
Methyl-2- (2-cyclohexyl-6- (3-cyclohexyl-2-hydroxy-5-methyl benzyl)-4-methyl phenoxy) acetate (2)
A stirred solution of 6,6`-methylenebis(2-cyclohexyl-4-methylphenol) (1) (100mg,0.239 mmole) in acetone (10 mL) was treated with potassium carbonate (165 mg, 1.2 mmole), and heated under reflux for 1 h. Methylchloroacetate (0.0313 mL, 0.389 mmole) was added and heating was continued for 4 h. The reaction mixture was cooled to room temperature, filtered and the filtrate was concentrated under reduced pressure. The dichloromethane solution (20 mL) of the resulting syrup was washed with water and dried over anhydrous sodium sulfate. It was filtered and the filtrate was evaporated to give yellowish oil.TLC showed a single spot of monomer 2. The product was purified by column chromatography to give the required product in 70% yield as white solid. m.p.120-122°C.H1 NMR (400 MHz, CDCl3) δ ppm: 6.9(s, 1H, OH), 6.8(d, ,jHH=11.2, 3H, Ar-H), 6.70(s, 1H, Ar-H), 4.44 (s, 2H, OCH2), 3.85(s, 3H, OCH3), 3.83(s, 2H, Ar-CH2-Ar), 2.96 (m, 1H, cyclohexyl-H), 2.78 (s, 1H, cyclohexyl-H, 2.23 (d, ,jHH=4Hz, 6H, Ar-CH3), 1.79 (m, 10H, cyclohexyl-H), 1.38 (m, 10H, cyclohexyl-H). IR (KBr, cm-1): 3420, 2923, 2850, 1755, 1471, 1207, 1055. Molecular ion peak (M+); 464, Major fragmentation peaks; 464, 446, 432, 291, 262, 203, 121, 83, 55, 41.HR-EIMS [M+], Calc. for C30H40O4; 464.2927, found 464.2938.
2-(2-cyclohexyl-6-(3-cyclohexyl-2-hydroxy-5- methylbenzyl)-4-methyl phenoxy) acetohydrazide (3)
A stirred solution ofmethyl-2-(2-cyclohexyl-6-(3-cyclohexyl-2-hydroxy-5-metyl benzyl)-4-methyl phenoxy)acetate (50mg, 0.0933 mmole) in ethanol(10 mL) was heated under reflux for 10 minutes. Hydrazine hydrate,(0.00521 mL ,0.01073 mmoles) was added and heating under reflux continued for 3 h. The reaction mixture was cooled to room temperature; the colorless liquid of reaction mixture was concentrated under reduced pressure, washed with hexane and dried to give the required product in 73% yield as a solid. m.p.138°C. H1 NMR (400 MHz, CDCl3) δ ppm : 7.8(s, 1H, NH), 6.91(s, 1H, Ar-H), 6.85(s, H, Ar-H), 6.80(s, 1H, Ar-H, 6.72(s, 1H, Ar-H), 5.25(s, 1H, OH), 4.34 (s, 2H, OCH2), 3.95(s, 2H, NH2), 3.81(s, 2H, Ar-CH2-Ar), 2.73 (m,2H, cyclohexyl-H), 2.22 (s, 6H, Ar-CH3), 1.82(m, 10H, cyclohexyl-H), 1.41(m, 10H, cyclohexyl-H). IR (KBr, cm-1): 3400, 3197, 2923,1691, 1473, 1211, 1060. HR-EIMS [M+], Calc. for C29H40N2O3; 464.3039, found 464.3043.
2-(2-cyclohexyl-6-(3-cyclohexyl-2-hydroxy-5-methylbenzyl)-4-methylphenoxy)-N'-(propan-2-ylidene) acetohydrazide (4)
The stirred solution of 2-(2-cyclohexyl-6-(3-cyclohexyl-2-hydroxy-5- methylbenzyl)-4-methyl phenoxy) acetohydrazide in acetone was reflux for 3 hours and then after cooling to room temprature the solvent was evaporated under reduced pressure and then the compound was recrystilzed in ethanol to give the required prouduct as in 90% yield. H1 NMR (400 MHz, CDCl3) δ ppm : 7.8(s, 1H, NH), 6.91(s, 1H, Ar-H), 6.85(s, H, Ar-H), 6.80(s, 1H, Ar-H, 6.72(s, 1H, Ar-H), 5.25(s, 1H, OH), 4.34 (s, 2H, OCH2), 3.81(s, 2H, Ar-CH2-Ar), 2.73 (m,2H, cyclohexyl-H), 2.22 (s, 6H, Ar-CH3), 1.85 (s, 6H, 2-NHN-CH3) 1.82(m, 10H, cyclohexyl-H), 1.41(m, 10H, cyclohexyl-H). HR-EIMS [M+], Calc. for C32H44N2O3; 504.3352, found 504.3349.
Dimethyl 2,2`-(6,6`-methylenebis(2-cyclohexyl-4-methyl-6,1 phenylene) bis(oxy)diacetate (5)
A stirred solution of 6,6`-methylenebis(2-cyclohexyl-4-methylphenol) (100mg,0.239 mmole) in acetone (10 mL) was treated with potassium carbonate (165 mg 1.2 mmole), and heated under reflux for 1 h. Methylchloroacetate (0.0627 mL, 0.778 mmole) was added and heating was continued for 10 hours. The reaction mixture was cooled to room temperature, filtered and the filtrate was concentrated under reduced pressure. The dichloromethane solution (20 mL) of the resulting syrup was washed with water and dried over anhydrous sodium sulphate. It was filtered and the filtrate was evaporated to give yellowish oil. TLC showed a mixture of monomer and dimer, both separated by column chromatography. The product was purified by column chromatograohy to give the desired product in 75% Yield as a white solid m.p:123-125°C. 1H NMR (300 MHz, CDCl3) δ, ( ppm ):6.8 (s, 2H, Ar-H), 6.633(s, 2H, Ar-H), 4.293 (s, 4H, COCH2) 3.984(s, 2H, Ar-CH2-Ar), 3.775 (s, 6H, OCH3), 2.873 (m, 2H, cyclohexyl-H), 2.199 (s, 6H, Ar-CH3), 1.787 (m, 10H, cyclohexyl-H), 1.377 (m, 10H, cyclohexyl-H).IR (KBr, cm-1): 2927, 2850, 1768, 1438, 1201, 1083. Molecular ion peak (M+) ; 536, Major fragmentation peaks; 536, 504, 453, 373, 275, 261, 215, 185, 147, 135, 105, 81, 55, 45. HR-EIMS [M+], Calc. for C33H44O6; 536.3138, found 536.3129.
2,2`-methylenebis (2-cyclohexyl-4-methyl-6,1phenylene))bis(oxy) diaceto hydrazide (6)
A stirred solution of Dimethyl 2,2`-(6,6`-methylenebis (2-cyclohexyl -4-methyl- 6,1- phenylene) )bis (oxy) diacetate (50mg, 0.0933 mmole) in ethanol(10 mL) was heated under reflux for 10 minutes. Hydrazine hydrate, (0.01042 mL ,0.2146 mmoles) was added and heating under reflux continued for 3 h. The reaction mixture was cooled to room temperature, the colorless liquid of reaction mixture was concentrated under reduced pressure, washed with hexane and dried to give the desired product in 85% yield as a white solid. m.p: 138 °C. 1H NMR (DMSO-d6; 400 MHz) δ ppm: 9.28 (s, 2H, 2 NH), 6.91(s, 2H, Ar-H), 6.59(s, 2H, Ar-H), 4.32(s, 4H, NH2) 4.07(s, 4H COCH2), 3.92(s, 2H, Ar-CH2- Ar), 2.81 (m, 2H, cyclohexyl-H), 2.15(s, 6H, Ar-CH3), 1.76(m, 10H, cyclohexyl-H), 1.36 (m, 10H, cyclohexyl-H). (KBr, cm-1): 3400, 3199, 2923, 2848, 1691, 1469, 1213, 1060. Molecular ion peak (M+); 536, Major fragmentation peaks; 536, 505, 464, 432, 392, 373, 243, 203, 190, 105, 44.HR-EIMS [M+], Calc. for C31H44N4O4; 536.3363, found 536.3364.
2,2`-(6-6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene)) bis (oxy) bis (N-(4-chlorobenzylidene) acetohydrazide) (7a)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl-4-methyl-6,1phenylene)) bis (oxy) diacetohydrazide (50mg, 0.0933 mmole) in ethanol(10 mL) was heated under reflux for 10 minutes. Para chloro benzaldehyde (39.3 mg, 0.28 mmole),3 drops of acetic acid was added and heating under reflux continued for 10 h.The content of the flask was cooled to room temprature. The liquid reaction mixture showed 3 spots on TLC in 8:2 hexane ethyl acetate solvent system. The product was purified by column chromatography, condensed and recrystallized in ethanol as solid with 70% yield. m.p:147-149°C. 1H NMR (DMSO-d6; 400 MHz) δ ppm:11.56 (s, 1H, NH), 11.5:11.46 (d, 1H, NH), 8.44:8.38:8.3(3s, 1H, NCH), 7.9:7.7 (d,jHH=5.6Hz, 1H, NCH) 7.68 (t, jHH=8.4,2H, Ar`-H), 7.38( d,jHH=8.4, 2H, Ar`-H), 7.48(d,jHH=8.8, 2H, Ar`-H),7.38(d,jHH=8.4, 2H, Ar`-H), 6.93(s, 1H, Ar-H), 6.89(s, 1H, Ar-H), 6.73(s, 1H, Ar-H), 6.79(s, 1H, Ar-H), 4.63 (d,jHH=12, 2H, OCH2), 4.24 (d,jHH=8.4, 2H, OCH2), 4 (d,jHH=4, 2H, Ar-CH2- Ar) 2.85 (s, 2H, cyclohexyl-H), 2.18(d,jHH=13.2, 2H, 6H, Ar-CH3), 1.71 (m, 10H, cyclohexyl-H), 1.34 (m, 10H, cyclohexyl-H).IR (KBr, cm-1): 3427, 2925, 2850, 1683, 1600, 1465, 1201, 1056. 823. Molecular ion peak (M+) ; 780, Major fragmentation peaks; 780, 627, 585, 490, 444, 432, 413, 397, 373, 307, 243, 229, 201, 167, 121, 105, 81, 55. HR-EIMS [M+], Calc. for C45H50Cl2N4O4; 780.3209, found 780.3140.
2,2`-(6,6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene)) bis (oxy) bis(N`benzylideneacetohydrazide) (7b)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl-4-methyl-6,1phenylene)) bis (oxy)diacetohydrazide (50mg, 0.0933 mmole) in ethanol(10 mL) was heated under reflux for 10 minutes. Benzaldehyde (0.0216 mL, 0.2146 mmole), three drops of acetic acid was added and heating under reflux continued for 9 h. The content of the flask was cooled to room temperature, filtered. On condensation, white precipitate was collected, washed with ethanol and recrystallized in ethanol as solid to give 60% yield. m.p:166°C.1H NMR (DMSO-d6; 400 MHz) δ ppm:11.51 (s, 1H, NH), 11.44 (d,jHH=12.4 Hz, 1H, NH), 8.47:8.41(2s, 1H, NCH), 7.94:7.90 (d,jHH=14 Hz, 1H, NCH) 7.66 (s, 2H, Ar`-H), 7.48( m, 8H, Ar`-H), 6.94:6.89(2s, 2H, Ar-H),6.76:6.670 (2s, 2H, Ar-H), 4.63:4.58 (2s, 2H, OCH2), 4.25:4.21 (2s, 2H, OCH2), 4 (d ,jHH=8.8, 2H, Ar-CH2-Ar) 2.88 (s, 2H, cyclohexyl-H), 2.18(d,jHH=15.2, 6H, Ar-CH3), 1.68 (m, 10H, cyclohexyl-H), 1.37 (m, 10H, cyclohexyl-H).IR (KBr, cm-1): 3406, 2923, 2850, 1689, 1470, 1203, 1056. Molecular ion peak (M+); 712, Major fragmentation peaks; 712, 593, 551, 534, 432, 373, 243, 201, 181, 161, 133, 119, 105, 81, 69, 55, 44. HR-EIMS [M+], Calc. for C45H52N4O4; 712.3989, found 712.3928.
2,2`-(6,6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene)) bis (oxy) bis(N`-(3-bromobenzylidene) acetohydrazide) (7c)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl-4-methyl-6,1phenylene)) bis (oxy)diacetohydrazide (50mg, 0.0933 mmole) in ethanol(10 mL) washeatedunderreflux for 20 minutes. 3-bromobenzaldehyde (0.0245 mL, 0.2146 mmole), 3 drops of acetic acid was added and heating under reflux continued for 9 h. The content of the flask was cooled to room temperature and filtered. White precipitate was collected, washed with ethanol and recrystallized from dichloromethane, ethanol in 75% yield as solid. m.p:213-215°C. 1H NMR (DMSO-d6; 400 MHz) δ ppm:11.64 (d,jHH=14.7Hz 1H, NH), 11.53 (s, 1H, NH), 8.81(d,jHH=14.1Hz, 1H, NCH), 7.85(s, 1H, NCH) 7.81 (s, H, Ar`-H), 7.69(m, 3H, Ar`-H), 7.53 (m, 2H, Ar`-H), 7.39(m, 2H, Ar`-H), 6.93(s, 1H, Ar-H), 6.88(d,jHH=6.9, 1H, Ar-H), 6.72(d,jHH=5.7, 2H, Ar-H), 4.60 (s, 2H, OCH2), 4.25 (d,jHH=8.1, 2H, OCH2), 3.99 (s, 2H, Ar-CH2-Ar) 2.82 (m, 2H, cyclohexyl-H), 2.18(s, 3H, Ar-CH3), 2.16 (d,jHH=4.8, 3H, Ar-CH3), 1.68 (m, 10H, cyclohexyl-H), 1.37 (m, 10H, cyclohexyl-H). IR (KBr, cm-1): 3427, 2923, 2850, 1693, 1544, 1447, 1288, 1207, 1068.Molecular ion peak (M+); 870, Major fragmentation peaks; 870, 673, 631, 490, 432, 373, 243, 229, 203, 182,102, 81, 55. HR-EIMS [M+], Calc. for C45H50Br2N4O4; 868.2199, found 868.2110.
2,2`-(6,6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene))bis (oxy) bisN`-(4-fluorobenzylidene) acetohydrazide) (7d)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl -4-methyl-6,1 phenylene)) bis (oxy)diacetohydrazide (50mg, 0.0933 mmole) in ethanol(10 mL) was heated under reflux for 20 minutes. Para fluorobenzaldehyde (0.0232 mL, 0.215 mmole), three drops of acetic acid was added and heating under reflux continued for 9 h. The content of the flask was cooled to room temperature and filtered. White precipitate was collected, washed with ethanol and recrystallized from dichloro methane, ethanol as solid in 65% yield. m.p: 211-213°C. 1H NMR (DMSO-d6; 400 MHz) δ ppm:11.50 (s, 1H, NH), 11.44 (d,jHH=15.6, 1H, NH), 8.45:8.39:(2s, 1H, NCH), 7.89(d,jHH=5.2Hz, 1H, NCH) 7.73 (m, 2H, Ar`-H), 7.55(m, 2H, Ar`-H), 7.24 (m, 2H, Ar`-H),7.18 (m, 2H, Ar`-H), 6.93(s, 1H, Ar-H), 6.89(s, 1H, Ar-H), 6.73(s, 1H, Ar-H), 6.69(s, 1H, Ar-H), 4.63 (d jHH=15.2, 2H, OCH2), 4.24 (d,jHH=9.2, 2H, OCH2), 4.01 (d,jHH=7.2, 2H, Ar- CH2-Ar) 2.85 (m, 2H, cyclohexyl-H), 2.18(s, 3H, Ar-CH3), 2.15 (s, 3H, Ar-CH3), 1.71 (m, 10H, cyclohexyl-H), 1.37 (m, 10H, cyclohexyl-H).IR (KBr, cm-1): 3421, 2925, 1689, 1234, 1028, 1002, 831.Molecular ion peak (M+); 748, Major fragmentation peaks; 748, 730, 690, 611, 569, 552, 444, 432, 414, 381, 373,243, 203, 190, 121, 94, 55. HREIMS [M+], Calc. for C45H50F2N4O4; 748.3800, found 748.4055.
2,2`-(6,6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene)) bis (oxy) bis(N`-(2-chlorobenzylidene) acetohydrazide) (7e)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl-4-methyl-6,1phenylene)) bis (oxy)diacetohydrazide (60mg, 0.1119 mmole) in ethanol(10 mL) was heated under reflux for 20 minutes.2 chlorobenzaldehyde (0.036 g , 0.2578 mmole),3 drops of acetic acid was added and heating under reflux continued for 12 h. The content of the flask was cooled to room temperature and filtered. On condensation, white precipitate was collected, washed with ethanol and recrystallized from dichloro methane, ethanol in 65% yield as solid. m.p:148-150°C.1H NMR (DMSO-d6; 400 MHz) δ ppm:11.71 (d,jHH=15.2Hz 1H, NH), 11.60 (s, 1H, NH), 8.84(d,jHH=13.6Hz, 1H, NCH), 7.28(d,jHH=4.8Hz, 1H, NCH) 7.96 (m, H, Ar`-H), 7.66(m, H, Ar`-H), 7.49 (m, 6H, Ar`-H), 6.94(s, 1H, Ar-H), 6.87(s, 1H, Ar-H), 6.83(s, 1H, Ar-H), 6.75(s, 1H, Ar-H), 4.63 (d jHH=11.2, 2H, OCH2), 4.26 (d,jHH=7.2, 2H, OCH2), 4.02 (m, 2H, Ar-CH2-Ar) 2.84 (m, 2H, cyclohexyl-H), 2.19(s, 3H, Ar-CH3), 2.14 (s, 3H, Ar-CH3), 1.71 (m, 10H, cyclohexyl-H), 1.34 (m, 10H, cyclohexyl-H).IR (KBr, cm-1): 3425, 2923, 2850, 1695, 1598, 1440, 1199, 1051.Molecular ion peak (M+) ; 781, Major fragmentation peaks;781, 700, 397, 315, 260, 203, 167. HR-EIMS [M+], Calc. for C45H50Cl2N4O4; 780.3209, found 780.3140.
2,2`-(6,6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene)) bis (oxy) bis(N`-(4-methylbenzylidene) acetohydrazide) (7f)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl -4-methyl-6,1 phenylene)) bis (oxy)diacetohydrazide (50mg, 0.0933 mmole) in ethanol(10 mL) was heated under reflux for 15 minutes.Para methylBenzaldehyde (0.025 mL , 0.217 mmole),3 drops of acetic acid was added and heating under reflux continued for 10 h. The content of the flask was cooled to room temperature. On condensation, yellowish precipitate was collected, washed with ethanol and recrystallized from dichloro methane, ethanolin 50% yield as solid. m.p:152°C. 1H NMR (DMSO-d6; 400 MHz) δ ppm: 11.45 (s, 1H, NH), 11.38 (d,jHH=5.7, 1H, NH), 8.42:8.35:(2s, 1H, NCH), 7.86(d,jHH=3.6Hz, 1H, NCH) 7.81 (d jHH=8.1,2H, Ar`-H), 7.56( d,jHH=6.9, 2H, Ar`-H), 7.42(m, 2H, Ar`-H), 7.23(d,jHH=7.2, 2H, Ar`-H), 6.93(s, 1H, Ar-H), 6.87(s, 1H, Ar-H), 6.76(s, 1H, Ar-H), 6.69(s, 1H, Ar-H), 4.61:4.55 (2s, 2H, OCH2), 4.24 (d,jHH=14.1, 2H, OCH2), 4.00 (d,jHH=6.3, 2H, Ar-CH2-Ar) 2.85 (m, 2H, cyclohexyl-H), 2.31(s, 3H, Ar`-CH3), 2.26 (s, 3H, Ar`-CH3), 2.18(d,jHH=2.1, 3H, Ar-CH3 ), 2.15(s, 3H, Ar-CH3), 1.67 (m, 10H, cyclohexyl-H), 1.30 (m, 10H, cyclohexyl-H). IR (KBr, cm-1): 3419, 2923, 2854, 2543, 1679, 1606, 1417, 1286, 1178, 958. Molecular ion peak (M+); 740, Major fragmentation peaks; 740, 722, 607, 565, 548, 490, 432, 414, 391, 373, 201, 135, 117, 105, 91, 81, 55, 44. HR-EIMS [M+], Calc. for C47H56N4O4; 740.4302, found 740.5016.
2,2`-(6,6`-methylenebis (2-cyclohexyl-4-methyl-6,1-phenylene)) bis (oxy) bis(N`-(4-(dimethylamino)benzylidene) acetohydrazide) (7g)
A stirred solution of 2,2`-methylenebis(2-cyclohexyl-4-methyl-6,1phenylene)) bis (oxy)diacetohydrazide (50mg, 0.0933 mmole) in ethanol (10 mL) was heated under reflux for 20 minutes. 4-(dimethylamino) benzaldehyde (0.0320g, 0.215 mmole), 3 drops of acetic acid was added and heating under reflux continued for 8 h. The content of the flask was cooled to room temperature and filtered. White precipitate was collected, washed with ethanol and recrystallized from ethanol in 60% yield as solid. m.p:253-255°C.1H NMR (DMSO-d6; 400 MHz) δ ppm:11.22 (s, 1H, NH), 11.15 (d,jHH=13.2, 1H, NH), 8.31:8.24:(2s, 1H, NCH), 7.77(d,jHH=4.8Hz, 1H, NCH) 7.48 (d jHH=8.4,2H, Ar`-H), 7.32:7.29:7.27( 3s, 2H, Ar`-H), 6.94(s, 1H, Ar-H), 6.90 d,jHH=8.4, 1H, Ar-H), 6.77 (s, 1H, Ar`-H), 6.70(m, 3H, Ar`-H), 6.62(m, 2H, Ar-H), 4.62:4.54 (2s, 2H, OCH2), 4.21:4.16 (2s, 2H, OCH2), 4.00 (t, 2H, Ar-CH2-Ar) 2.95 (d,jHH=3.2, 6H, NCH3), 2.90 (d,jHH=10.46, 6H, NCH3), 2.72(m, 2H, , cyclohexyl-H), 2.19(d,jHH=4, 3H, Ar-CH3), 2.15(s, 3H, Ar-CH3), 1.69 (m, 10H, cyclohexyl-H), 1.66 (m, 10H, cyclohexyl-H). IR (KBr, cm-1): 3431, 2923, 2854, 1678, 1604, 1431, 1236, 1174. Molecular ion peak (M+) 799, Major fragmentation peaks; 799, 406, 204, 176, 163, 147. HR-EIMS [M+], Calc. for C49H62N6O4; 798.4833, found 798.4830.
3D-pharmacophore-based ligand alignment, in silico ADMET screening and molecular docking study
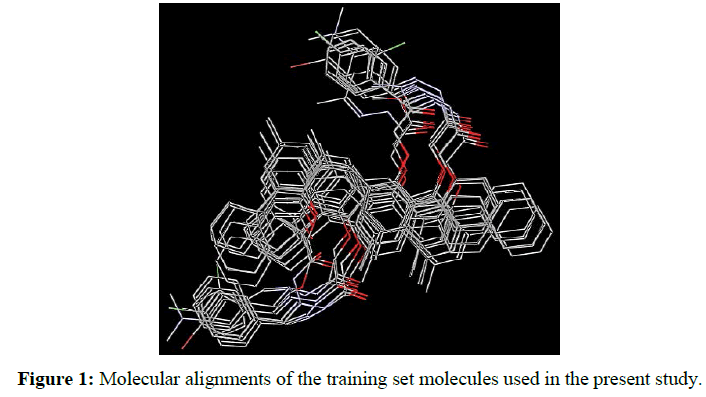
3D-pharmacophore-based ligand alignment: The alignment rule was defined by a chemical function-mapping method and therefore based on a geometric fit of the chemical functions of the molecules to the chemical features of the pharmacophore. All generated conformations were developed in Discovery Studio (DS) 2.5 software (Accelrys software Inc.) using its accurate pattern-matching 3D alignment algorithm. The compounds were converted to 3D structure and minimized its MMFF94 energy with 200 iteration limit and energy threshold value of 15 kcal/mol above the global energy minimum until a local energy minimum was reached [37,38]. This algorithm allows all internal coordinates to vary and energetically analyzes all rings and chains. The conformation with highest fit value (i.e., best fitting the pharmacophore) was assumed as the bioactive conformation for each compound. The final aligned molecules (Figure 1) were used to generate the pharmacophore model of this class of compounds.
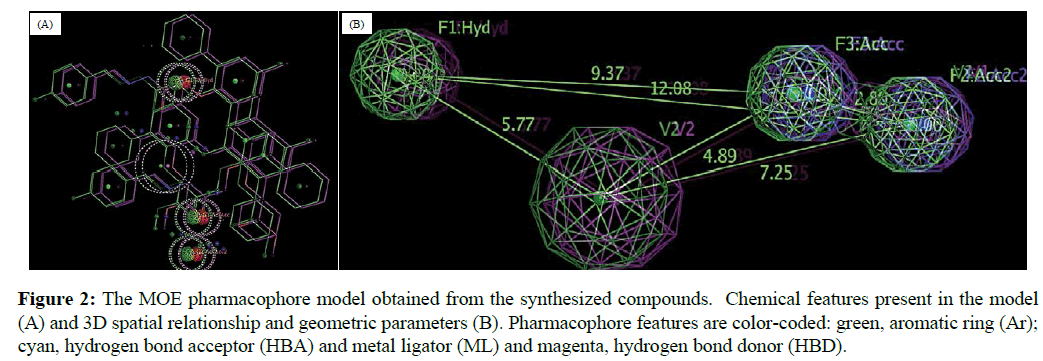
The pharmacophore models were created for the aligned molecules using common feature pharmacophore generation in DS software. HipHop module of Catalyst which was popularly known for common feature pharmacophore generation is available in this program as Common Feature Pharmacophore Generation protocol [39]. The default value of 2.97Å for the Minimum Inter-feature Distance was changed to 2 during pharmacophore generation. Pharmacophore sites of a ligand are represented in the 3D space by a set of points. Other parameters, like the maximum number of 255 conformers and an energy threshold value of 20 kcal/mol above the global energy minimum, were chosen during conformation generation. During the pharmacophore hypothesis generation, pharmacophoric features such as hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), hydrophobic feature (Hyd), and aromatic region (Ar) were used to create reliable pharmacophore sites for the energy calculated ligands for our experimental results. Hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), hydrophobic feature (Hyd), and aromatic region (Ar) chemical features were chosen on the basis of the chemical charectersitics of the compounds using Feature Mapping protocol available in DS. The standard functions can be effectively used in place of the metal bonding chemical function, as there are no special structures available for metal bonding in the DS feature dictionary [40,41].
In silico ADMET screening
The synthesized compounds were submitted to an in silico ADMET screening, using the DS 2.5 software to analyze their overall drug score and toxicity risks, compared to the available drugs used for chemotherapy [42]. The ADMET properties including aqueous solubility, blood brain barrier (BBB), plasma protein binding, CYP2D6 binding, intestinal absorptionand hepatotoxicity were evaluated for these molecules within human. In addition, AlogP98 and PSA_2D were used in plotting the confidence ellipses. The models used to predict the ADMET properties in this protocol are derived from a variety of experimental data sources and are catalogued in the product documentation.
Screening for pharmacokinetics and drug-likeness
Pharmacokinetics and drug-likeness prediction for the synthesized compounds were also performed by online tool SwissADME [43] of Swiss Institute of Bioinformatics (https://www.sib.swiss) was used to evaluate individual ADME behaviors of those compounds [44]. 2D structural models were drawn in ChemBioDraw Ultra version 15.0 (Cambridge Software) and SMILES of each compound was translated into molfile by online SMILES translator and structure file generator found in Online tool SwissADME. The analysis task was done to check whether those compounds were inhibitor of isoforms of Cytochrome P450 (CYP) family such as CYP1A2 and CYP2D6. In addition, pharmacokinetics (such as gastro intestinal absorption, P-glycoprotein and Blood brain barrier) and drug-likeness prediction such as Lipinski, Ghose and Veber rules and bioavailability score [45-47]. The Lipinski, Ghose and Veber rules were applied to assess druglikeness to predict whether a compound is likely to be a bioactive according to some important parameters such as molecular weight, LogP, number of HPA and HBD. The Swiss ADME tool used vector machine algorithm (SVM) [48] with fastidiously cleaned large datasets of known inhibitors/non-inhibitors as well as substrates/non-substrates.
Principal component analysis
Principal component analysis (PCA) in DS 2.5 software (Accelrys, San Diego, CA) was used to predict models showed the molecular descriptor including AlogP; molecular weight; number of hydrogen bond donors (HBD); number of hydrogen bond acceptors (HBA); number of rotatable; number of rings; number of aromatic rings and molecular polar surface area (PSA).
Molecular docking
A Molecular Operating Environment (MOE 2014.13, Chemical Computing Group Inc, Montreal, Quebec, Canada, https://www.chemcomp.com) software was used for molecular docking of the synthesized compounds on the target proteins [49]. The compounds were converted to 3D structure and minimized its MMFF94 energy with 200 iteration limit and energy threshold value of 15 kcal/mol above the global energy minimum until a local energy minimum was reached [37,38]. This algorithm allows all internal coordinates to vary and energetically analyses all rings and chains. A crystal structure of CYP1A2 (cytochrome P450 family 1 subfamily A member 2, PDB: 2HI4) and CYP2D6 (cytochrome P450 family 2 subfamily D member 6, PDB: 5TFT) as the selected inhibitors suggested from ADMET and SwissADME prediction were obtained at 3.58 Å and downloaded from the protein data bank (PDB)(https://www.rcsb.org/pdb/home/home.do). These structures were protonated in MOE software [49]. The active sites were defined with a radius of 4 Å around the bound inhibitor in the crystal structure of each protein. The triangle matching algorithm of MOE software was selected to dock selected identified compounds to the active site of the protein. The scoring function shall conform to the following parameters: (1) specify the ASE score to rank the poses exited by the placement step; (2) specifying the Force field refinement to release the poses and (3) specifying affinity ΔG scoring to classify the poses using the refinement step [38,50,51]. The free energy of the binding was calculated from the contributions of the hydrophobic, ionic, hydrogenated and van der Waals interactions between protein and ligand, intramolecular hydrogen bonds and ligand strains. We have observed that the docking poses were classified by the calculation of the free binding energy in the S field. Once the docked enzyme-ligand complexes were completed, an analysis of the binding sites was performed in order to create a ligand interaction 2D diagram for each compound.
Results and Discussion
Synthesis of compounds
Methyl-2-(2-cyclohexyl-6- (3-cyclohexyl-2-hydroxy-5-methylbenzyl)-4-methylphenoxy) acetate (2) was prepared by treating 6,6`-methylenebis(2-cyclohexyl-4-methylphenol) (1) with chloromethyl acetate (1 equivalent ) and potassium carbonate using acetone as solvent under reflux condition. This has indicated the possible selectivity in the carbomethoxymethylation process to give good yield (Scheme 1). On the other hand, similar alkylation of 6,6`-methylenebis(2-cyclohexyl-4-methylphenol) (1) using two equivalents of chloromethyl acetate gave dimethyl- 2,2`-(6,6`-methylenebis(2-cyclohexyl-4-methyl6,1phenylene)bis(oxy)diacetate (5).
Pharmacophore elucidation based on ligand alignment
In this study, the final aligned molecules as shown in Figure 1 were used to generate the pharmacophore model of this class of compounds. The automatic construction and visualization of the 3D pharmacophore models from the structural data of mono- and di-carbmethoxylated 6,6'-methylenebis(2-cyclohexyl-4-methylphenol) and their hydrazides and hydrazones was created by DS 2.5 software and the results are shown in Figure 2. The investigated pharmacophoric features included HBA, HBD, Hyd, and Ar. The basic structural requirements identified by DS software dependably consist of HBA, HBD, or both for metal binding.
Figure 2: The MOE pharmacophore model obtained from the synthesized compounds. Chemical features present in the model (A) and 3D spatial relationship and geometric parameters (B). Pharmacophore features are color-coded: green, aromatic ring (Ar); cyan, hydrogen bond acceptor (HBA) and metal ligator (ML) and magenta, hydrogen bond donor (HBD).
In silico ADMET evaluation and comparison with current drugs
In this study, we submitted the synthesized compounds to in silico ADMET screening, using the DS 2.5 software, to predict their overall absorption, distribution, metabolism, excretion, and toxicitytoxicity risks. In the in silico evaluation, the analysis of different descriptors (calculated octanol/water partition coefficient, molecular weight, molecular volume, and number of hydrogen bond donor and acceptor groups) of all compounds revealed that they are highly hydrophobic to penetrate the biological membranes, as determined by the Lipinski “rule-of-five” (cLogP˂5, MW>500, HBD<5 and HBA<10) [45,52]. The synthesized products have cLogP˃5 (ranged from 5.85 to 11.87) as shown in Table 1. Therefore, these results suggest that most of these compounds have a low theoretical oral bioavailability according to the Lipinski “rule-of-five”.
| Compound | In silico ADMET properties | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BBB level | Absorption level | Water solubility | Solubility level | Hepatotoxicity | Hepatotoxicity probability | CYP2D6 | CYP2D6 probability | PPB level | AlogP98 | PSA 2D | |
| (log s) | |||||||||||
| 1 | 4 | 3 | -8.148 | 0 | 0 | 0.39 | 0 | 0.277 | 2 | 8.68 | 41.63 |
| 2 | 4 | 3 | -8.463 | 0 | 0 | 0.483 | 0 | 0.415 | 2 | 8.61 | 55.98 |
| 3 | 4 | 3 | -7.701 | 1 | 1 | 0.536 | 0 | 0.316 | 2 | 7.91 | 71.18 |
| 4 | 4 | 3 | -7.637 | 1 | 0 | 0.476 | 0 | 0.415 | 2 | 7.26 | 86.4 |
| 5 | 4 | 3 | -8.322 | 0 | 0 | 0.39 | 0 | 0.435 | 2 | 8.54 | 70.32 |
| 6 | 4 | 2 | -7.683 | 1 | 0 | 0.403 | 0 | 0.316 | 2 | 5.85 | 131.16 |
| 7a | 4 | 3 | -9.213 | 0 | 0 | 0.456 | 0 | 0.089 | 2 | 11.87 | 100.73 |
| 7b | 4 | 3 | -8.196 | 0 | 0 | 0.476 | 0 | 0.108 | 2 | 10.54 | 100.73 |
| 7c | 4 | 3 | -9.407 | 0 | 1 | 0.536 | 0 | 0.128 | 2 | 12.03 | 100.73 |
| 7d | 4 | 3 | -8.412 | 0 | 0 | 0.483 | 0 | 0.089 | 2 | 10.95 | 100.73 |
| 7e | 4 | 3 | -9.308 | 0 | 1 | 0.529 | 0 | 0.118 | 2 | 11.87 | 100.73 |
| 7f | 4 | 3 | -8.564 | 0 | 0 | 0.483 | 0 | 0.089 | 2 | 11.51 | 100.73 |
| 7g | 4 | 3 | -6.567 | 1 | 1 | 0.576 | 0 | 0.069 | 2 | 10.86 | 107.43 |
Table 1: In silico ADMET properties of the synthesized compounds.
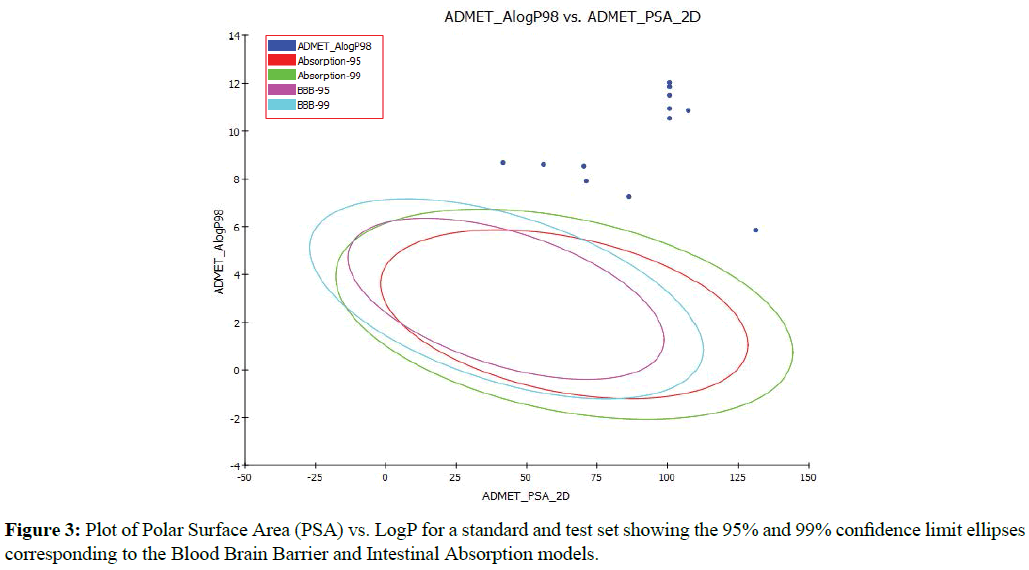
An ADMET model was also generated to predict the human intestinal absorption (HIA) after oral administration of the inhibitors tested. The model includes 95% and 99% confidence ellipses in the ADMET_PSA_2D and ADMET_ AlogP98 plane as shown in Figure 3. There are four prediction levels for the absorption of compounds as good (0), moderate (1), poor (2) and very poor (3). These levels are defined by the 95% (red line) and 99% (green line) confidence ellipsoids (Figure 3). The upper limit of PSA_2D value for the 95% confidence ellipsoid is at 131.16 for compound 6, while the lower value is 41.63 for compound 1 (Table 1). All of these values for 95 and 99% confidence levels are higher than that standard range shown in Figure 3, which confirm that these compounds are very poor in the absorption through human intestinal (ADMET_AlogP98˃5) as shown in Figure 3 and Table 1. Based on the in silico ADMET analysis, it was found that the test compounds did not accomplish the ADMET descriptors criteria at the optimal level (Table 1 and Figure 3).
Figure 3: Plot of Polar Surface Area (PSA) vs. LogP for a standard and test set showing the 95% and 99% confidence limit ellipses corresponding to the Blood Brain Barrier and Intestinal Absorption models.
Pharmacokinetics and drug-likeness prediction by SwissADME
The pharmacokinetic properties and drug-likeness prediction of the 13 compounds were performed by SwissADME online version and the data are shown in Table 2. According to the pharmacokinetic properties, all compound showed low Gastro intestinal absorption except compound 3 that has high absorption. All compounds have no BBB permeability however, most of them showed inhibition to Cytochrome P450 isomers (CYP1A2 and CYP2D6). The drug-likeness prediction was also conducted depending on the selected Lipinski, Ghose and Veber rules and bioavailability score.
| Compound | Pharmacokinetics | Drug-likeness | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GI absorption | BBB permeant | P-gp | CYP1A2 inhibitor | CYP2D6 inhibitor | Log Kp (skin permeation), cm/s | Lipinski | Ghose | Veber | Bioavailability Score | ||
| 1 | Low | No | Yes | No | Yes | -2.35 | Yes | No; 1 violation | Yes | 0.55 | |
| 2 | Low | No | Yes | No | Yes | -2.62 | Yes | No; 3 violations | Yes | 0.55 | |
| 3 | High | No | Yes | No | Yes | -3.55 | Yes | No; 3 violations | Yes | 0.55 | |
| 4 | Low | No | Yes | No | Yes | -3.13 | No; 2 violations | No; 4 violations | Yes | 0.17 | |
| 5 | Low | No | Yes | No | Yes | -2.88 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 6 | Low | No | No | No | No | -4.76 | Yes | No; 3 violations | No; 1 violation | 0.55 | |
| 7a | Low | No | Yes | Yes | No | -1.98 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 7b | Low | No | Yes | Yes | No | -2.46 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 7c | Low | No | Yes | Yes | No | -2.44 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 7d | Low | No | Yes | Yes | No | -2.53 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 7e | Low | No | Yes | Yes | No | -1.98 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 7f | Low | No | Yes | Yes | No | -2.11 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
| 7g | Low | No | Yes | No | No | -2.8 | No; 2 violations | No; 4 violations | No; 1 violation | 0.17 | |
GI: Gastro Intestinal; P-gp: P-glycoprotein; BBB: Blood Brain Barrier; CYP1A2: Cytochrome P450 family 1 subfamily A member 2 (PDB: 2HI4); CYP2D6: Cytochrome P450 family 2 subfamily D member 6 (PDB: 5TFT)
Table 2: Pharmacokinetics and drug-likeness prediction for the synthesized compounds by SwissADME.
The Lipinski (Pfizer) filter is the pioneer rule-of-five [53]. The Lipinski's Rule of Five states that the absorption or permeation of a molecule is more likely when the molecular weight is under 500 g/mol, the value of log P is lower than 5, and the molecule has utmost 5 H-donor and 10 H-acceptor atoms. Ghose filter (Amgen) [46] defines drug-likeness constraints as follows: calculated log P is between -0.4 and 5.6, MW is between 160 and 480, molar refractivity is between 40 and 130, and the total number of atoms is between 20 and 70. Veber (GSK) [47], rule defines druglikeness constraints as Rotatable bond count ≤ 10 and polar surface area (PSA) ≤ 140. The Bioavailability score was implemented without changes from Martin et al., and it is similar but seeks to predict the probability of a compound to have at least 10% oral bioavailability in rat or measurable Caco-2 permeability [54]. Screening process with Lipinski Rule of Five showed that there were only four compounds (1, 2, 3 and 6) meet the criteria of druglikeness assessment however, 9 compounds were rejected with two violations (Table 2).According to the screening process with Ghose rules showed that all compounds were rejected with one, three or four violations (Table 2). However, the screening process with Veber rules, compounds 1-4 meet the criteria of drug likeness assessment however, the other nine compounds (5-7 g) were rejected with two violations (Table 2).
Prediction of principal component models
Principal Component Analysis (PCA) was conducted on the molecular descriptors of the input molecules by DS 2.5 software. According to the physicochemical characterizes (ALogP, molecular weight, HBA, HBD, rotatable bonds, rings, aromatic ring and polar surface area) of the synthesized molecules, three models were generated. The equation coefficients of the PCA Temp models are shown in Table 3. PCA is an orthogonal linear transformation technique that transforms the data into a new coordinate system such that the variance of the data is maximized on the first coordinate (called the first principal component), the rest of the variance maximized on the second coordinate, and so on. The data in Table 3 showed that the high coefficients values were obtained with ring and ring aromatics parameters for model PC1. HBD and Polar Surface Area have the highest coefficient values in model PC2 however; HBD, ring and ring aromatics parameters have the highest coefficient values in model PC3.
| Equation term | Coefficient | ||
|---|---|---|---|
| Model PC1 | Model PC2 | Model PC3 | |
| Constant | -9.27223 | -5.22024 | -1.04444 |
| ALogP | 0.203272 | -0.124654 | 0.140863 |
| Molecular weight | 0.0027689 | 0.00059164 | 0.00033117 |
| HBD | -0.066076 | 0.702766 | 0.659005 |
| HBA | 0.238346 | 0.185972 | -0.389919 |
| Rotatable bonds | 0.117164 | 0.0330708 | -0.0945932 |
| Rings | 0.425499 | 0.0233489 | 0.257113 |
| Aromatic rings | 0.425499 | 0.0233489 | 0.257113 |
| Polar surface area | -3.08878 | 21.3919 | -5.34568 |
Table 3: Equation coefficients of the PCA models.
Molecular docking
In the present study, the automated docking was used to determine the orientation of mono- and di-carbmethoxylated 6,6'-methylenebis(2-cyclohexyl-4-methylphenol) and their hydrazides and hydrazones bound in the active site of the target enzymes of CYP1A2 (cytochrome P450 family 1 subfamily A member 2, PDB: 2HI4) and CYP2D6 (cytochrome P450 family 2 subfamily D member 6, PDB: 5TFT) as the selected inhibitors suggested from ADMET and SwissADME prediction (Table 4). The structures of the compounds as well as the enzymes were kept flexible to obtain different binding conformations and the best-docked complex obtained from it was analysed in detail. The molecular docking protocol was validated by extracting the native specific ligand of each enzyme from the binding site and then docking the conformations of the compounds to the binding site with less than 4ºA which validating the reliability and reproducibility of the docking procedure.
| Compound | CYP1A2 | CYP2D6 | ||
|---|---|---|---|---|
| Docking score (ΔG, kcal/mol) | H-bonds | Docking score (ΔG, kcal/mol) | H-bonds | |
| 1 | -11.49 | - | -8.83 | Ser304 |
| 2 | -11.36 | Ile459, Thr124 | -7.13 | Phe120 |
| 3 | -12.46 | Arg108, Ile335 | -7.29 | Glu215, Thr375 |
| 4 | -11.56 | Thr124 | -6.05 | Gly373, Thr309 |
| 5 | -11.91 | Arg105, Ile256, Gly480 | -5.3 | Arg221, Gln244, Val308 |
| 6 | -9.42 | Arg108, Gly316 | -5.58 | Cys443, Pro435 |
| 7a | -5.65 | Arg105, Gln411 | 1.09 | Arg101, Ala305 |
| 7b | -5.45 | Phe225 | 0.82 | Cys443, Ser204 |
| 7c | -4.62 | Phe451, Thr124 | 2.01 | Cys443, Gly306, Glu216 |
| 7d | -8.05 | Leu455, Phe225, Phe451 | 1.21 | Cys443, Phe120 |
| 7e | -6.37 | Ala284, Ile255 | 2.31 | Ala442, Leu302, Thr309 |
| 7f | -2.25 | Gly318, Gly452 | 1.12 | Glu215 |
| 7g | -4.02 | Phe225, Phe451 | 4.25 | Asp301 |
CYP1A2: Cytochrome P450 family 1 subfamily A member 2 (PDB: 2HI4); CYP2D6 (cytochrome P450 family 2 subfamily D member 6, PDB: 5TFT)
Table 4: Molecular docking, binding scores and binding reactions of compounds 1-13 within the active sites of CYP1A2 and CYP2D6. Residues/water molecules participating in hydrogen bonds and close van der Waals contacts (<4 Å) with the inhibitors are shown.
The results of the molecular docking have been analysed based on the docking score (ΔG, kcal/mol), hydrogen bonds and close van der Waals contacts and the data are shown in Table 4. The data show the intermolecular interaction energy values obtained from the docking calculation. Analysis of the docking poses showed that the thirteen products exhibited low to high binding affinity towards the sites of the target enzymes with docking energy ranging from -2.25 to -12.46 and from -8.83 to 4.25 kcal/mol for CYP1A2 and CYP2D6, respectively .
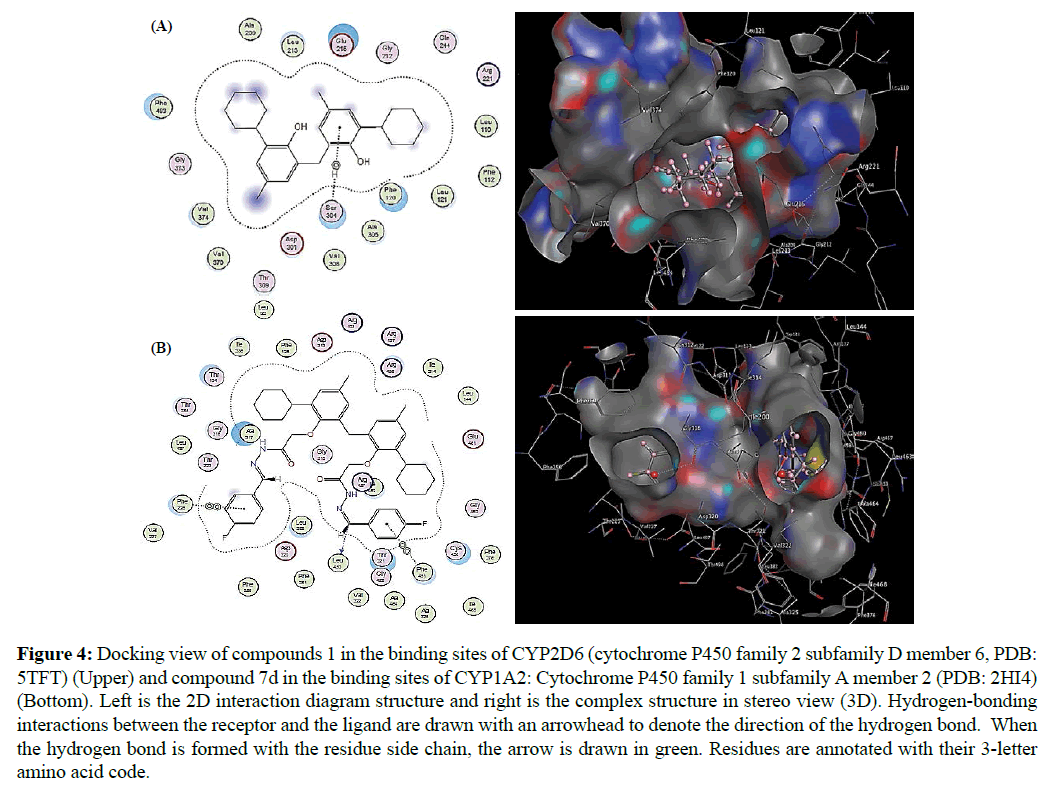
The docking of ligand molecules with enzymes reveals that all the derivatives are exhibiting the bonding with two or the other amino acids in the active pockets which is showed in Figure 4. From the resulting docking structures, it is clear that all products are attached to the active site of the target enzymes, subsequent in various close contacts with the amino acid residues that border the active site. Interaction analysis of the residues could also provide an elucidation of the difference observed in the binding affinity for these molecules. Compound 1 that showed the highest docking score (-8.83 kcal/mol) among the compounds that predicted an inhibition of CYP2D6 4 by SwissADME is stabilized within the active sites of the enzyme through extensive van der Waals contacts with many amino acids and hydrogen bond with Ser304 (Figure 4A). However, compound 7d that showed the highest docking score (-8.05 kcal/ mol) among the compounds that predicted an inhibition of CYP1A2 by SwissADME link to the active sites of the enzyme through extensive van der Waals contacts with many amino acids and hydrogen bond with Leu455, Phe225 and Phe451 (Figure 4B).
Figure 4: Docking view of compounds 1 in the binding sites of CYP2D6 (cytochrome P450 family 2 subfamily D member 6, PDB: 5TFT) (Upper) and compound 7d in the binding sites of CYP1A2: Cytochrome P450 family 1 subfamily A member 2 (PDB: 2HI4) (Bottom). Left is the 2D interaction diagram structure and right is the complex structure in stereo view (3D). Hydrogen-bonding interactions between the receptor and the ligand are drawn with an arrowhead to denote the direction of the hydrogen bond. When the hydrogen bond is formed with the residue side chain, the arrow is drawn in green. Residues are annotated with their 3-letter amino acid code.
Conclusion
Thirteen compounds of mono- and Di-carbmethoxylated 6,6'-Methylenebis(2-cyclohexyl-4-methylphenol) and their Hydrazides and Hydrazones were synthesized and their structures were determined using X-ray crystallography and spectroscopic techniques. Pharmacophore elucidation of the compounds was performed based on ligand alignment and the in silico ADMET analysis was done to predict the ADMET properties of the test compounds. Screening process with drug-likeness rules showed that there were few compounds meet the criteria of drug likeness however; the other products did not meet the criteria. In addition, molecular docking studies revealed that the compounds exhibited low to high binding affinity towards the sites of target enzymes of cytochrome P450 isomers (CYP1A2 and CYP2D6) with docking energy ranging from -12.46 to 4.25 kcal/mol.
Acknowledgments
We appreciate the partial support of the Higher Education Commission of Pakistan to perform this study through the Project No. 20-697/R&D/06/38.
References
- Ding XB, Yan X (2006) Study of piezoâ€ÂÂdamping properties of cpe/zkf/vgcf composites. J Appl Polym Sci 102: 3181-3185.
- Takemura H, Takahashi A, Suga H, Fukuda M, Iwanaga T (2011) Synthesis of azacalixarenes through dihydrobenzoxazine derivatives of phenols. Eur J Org Chem 31: 71-77.
- Biali SE, Böhmer V, Brenn J, Frings M, Thondorf I, et al. (1997) Conformation, inversion barrier, and solvent-induced conformational shift in exo-and endo/exo-calix arenes. J Org Chem 62: 8350-8360.
- Takeuchi D, Nakamura T, Aida T (2000) Bulky titanium bis (phenolate) complexes as novel initiators for living anionic polymerization of ε-caprolactone. Macromolecules 33: 725-729.
- Nifant'ev E, Teleshev A, Zhdanov A (2002) Cyclophosphites from oligomethylenephenols. Russ J Gen Chem 72: 903-908.
- Yu XF, Wang ZX (2013) Dinuclear aluminum complexes supported by amino-or imino-phenolate ligands: Synthesis, structures, and ring-opening polymerization catalysis of rac-lactide. Dalton Trans 42: 3860-3868.
- Ito K, Ito T, Takasawa T, Ohba Y (2002) Letters in organic chemistry anion receptors based on acyclic phenol-formaldehyde oligomers bearing thiourea groups. Lett Org Chem 3: 260-266.
- Woeste M, Steller J, Hofmann E, Kidd T, Patel R, et al. (2013) Structural requirements for inhibitory effects of bisphenols on the activity of the sarco/endoplasmic reticulum calcium atpase. Bioorg Med Chem 21: 3927-3933.
- Sakamoto T, Gemmell MA (1975) Studies on effects of drugs upon protoscoleces of echinococcus granulosus in vitro: I scolicidal effect of salicylanilide and bisphenol derivatives against ech1nococcus granulosus in-vitro. Jpn J Vet Res 23: 81-94.
- Ragavendran JV, Sriram D, Patel SK, Reddy IV, Bharathwajan N, et al. (2007) Design and synthesis of anticonvulsants from a combined phthalimide–gaba–anilide and hydrazone pharmacophore. Eur J Org Chem 42: 146-151.
- Ergenç N, Günay NS, Demirdamar R (1998) Synthesis and antidepressant evaluation of new 3-phenyl-5-sulfonamidoindole derivatives. Eur J Med Chem 33: 143-148.
- Todeschini AR, Miranda ALP, Silva KCM, Parrini SC, Barreiro EJ (1998) Synthesis and evaluation of analgesic, antiinflammatory and antiplatelet properties of new 2-pyridylarylhydrazone derivatives. Eur J Med Chem 33: 189-199.
- Radwan MA, Ragab EA, Sabry NM, El-Shenawy SM (2007) Synthesis and biological evaluation of new 3-substituted indole derivatives as potential anti-inflammatory and analgesic agents. Bioorg Med Chem 15: 3832-3841.
- Gemma S, Kukreja G, Fattorusso C, Persico M, Romano MP, et al. (2006) Synthesis of n1-arylidene-n2-quinolyl-and n2-acrydinylhydrazones as potent antimalarial agents active against CQ-resistant P. Falciparum strains. Bioorg Med Chem Lett 16: 5384-5388.
- Sahu NK, Sharma M, Mourya V, Kohli D (2012) QSAR study of some substituted 4-quinolinyl and 9-acridinyl hydrazones as antimalarial agents. Acta Pol Pharmaceutica 69: 1153-1165.
- Bijev A (2006) New heterocyclic hydrazones in the search for antitubercular agents: Synthesis and in vitro evaluations. Lett Drug Des Discov 3: 506-512.
- Nayyar A, Monga V, Malde A, Coutinho E, Jain R (2007) Synthesis, anti-tuberculosis activity, and 3d-qsar study of 4-(adamantan-1-yl)-2-substituted quinolines. Bioorg Med Chem 15: 626-640.
- Gürsoy E, Güzeldemirci NU (2007) Synthesis and primary cytotoxicity evaluation of new imidazo [2, 1-b] thiazole derivatives. Eur J Med Chem 42: 320-326.
- Sztanke K, Tuzimski T, Rzymowska J, Pasternak K, Kandefer SM (2008) Synthesis, determination of the lipophilicity, anticancer and antimicrobial properties of some fused 1, 2, 4-triazole derivatives. Eur J Med Chem 43: 404-419.
- Masunari A, Tavares CA (2007) New class of nifuroxazide analogues: Synthesis of 5-nitrothiophene derivatives with antimicrobial activity against multidrug-resistant staphylococcus aureus. Bioorg Med Chem 15: 4229-4236.
- Loncle C, Brunel JM, Vidal N, Dherbomez M, Letourneux Y (2004) Synthesis and antifungal activity of cholesterol-hydrazone derivatives. Eur J Med Chem 39: 1067-1071.
- Küçükgüzel SG, Mazi A, Sahin F, Öztürk S, Stables J (2003) Synthesis and biological activities of diflunisal hydrazide–hydrazones. Eur J Med Chem 38: 1005-1013.
- Vicini P, Zani F, Cozzini P, Doytchinova I (2002) Hydrazones of 1, 2-benzisothiazole hydrazides: Synthesis, antimicrobial activity and QSAR investigations. Eur J Med Chem 37: 553-564.
- Leite ACL, Lima RS, Moreira DRM, Cardoso MVO, Brito ACG, et al. (2006) Synthesis, docking, and in vitro activity of thiosemicarbazones, aminoacyl-thiosemicarbazides and acyl-thiazolidones against trypanosoma cruzi. Bioorg Med Chem 14: 3749-3757.
- Al-Mawsawi LQ, Dayam R, Taheri L, Witvrouw M, Debyser Z, et al. (2007) Discovery of novel non-cytotoxic salicylhydrazide containing hiv-1 integrase inhibitors. Bioorg Med Chem lett 17: 6472-6475.
- Abdel AMT, El Sayed WA, El Ashry ESH (2006) Synthesis and antiviral evaluation of some sugar arylglycinoylhydrazones and their oxadiazoline derivatives. Archiv Der Pharmazie 339: 656-663.
- Ashry ES, Ud DZ, Soomro ZH, Rahman W, Shah MR, et al. (2014) Synthesis, Characterization and Biological Evaluation Against InfluenzaVirus Agonists of (N'E,N'"E)-2,2'-[[1,1'-Biphenyl]-4,4'-dihylbis(oxy)]bis(N'arylmethyleneacetohydrazides). Lett Org 11: 168-173.
- Dearden JC (2006) In silico prediction of aqueous solubility. Exp Op Drug Disc 1: 31-52.
- Lombardo F, Gifford E, Shalaeva MY (2003) In silico adme prediction: Data, models, facts and myths. Mini Rev Med Chem 3: 861-875.
- Kulkarni S, Zhu J, Blechinger S (2005) In silico techniques for the study and prediction of xenobiotic metabolism: A review. Xenobiotica 35: 955-973.
- Gola J, Obrezanova O, Champness E, Segall M (2006) Admet property prediction: The state of the art and current challenges. Mol Inform 25: 1172-1180.
- Johnson SR, Zheng W (2006) Recent progress in the computational prediction of aqueous solubility and absorption. AAPS J 8: 27-40.
- Dearden JC (2003) In silico prediction of drug toxicity. J Comput Aid Mol Des 17: 119-127.
- Lewis DF, Ito Y, Goldfarb PS (2005) Cytochrome p450 structures and their substrate interactions. Drug Dev Res 66: 19-24.
- Graaf C, Vermeulen NP, Feenstra KA (2005) Cytochrome p450 in silico: An integrative modeling approach. J Med Chem 48: 2725-2755.
- Dearden JC (2007) In silico prediction of admet properties: How far have we come? Expert Opin Drug Metab Toxicol 3: 635-639.
- Halgren TA (1999) Mmff vi. Mmff94s option for energy minimization studies. J Comput Chem 20: 720-729.
- Vilar S, Cozza G, Moro S (2008) Medicinal chemistry and the molecular operating environment (moe): Application of qsar and molecular docking to drug discovery. Curr Top Med Chem 8: 1555-1572.
- Thangapandian S, John S, Sakkiah S, Lee KW (2010) Ligand and structure based pharmacophore modeling to facilitate novel histone deacetylase 8 inhibitor design. Eur J Med Chem 45: 4409-4417.
- Zhu Y, Li HF, Lu S, Zheng YX, Wu Z, et al. (2010) Investigation on the isoform selectivity of histone deacetylase inhibitors using chemical feature based pharmacophore and docking approaches. Eur J Med Chem 45: 1777-1791.
- Yang SY (2010) Pharmacophore modeling and applications in drug discovery: Challenges and recent advances. Drug Discov Today 15: 444-450.
- Discovery Studio. 2.1 (2008) San Diego, CA, USA.
- Zoete V, Daina A, Bovigny C, Michielin O (2016) SwissSimilarity: A Web tool for low to ultra high throughput ligand-based virtual screening. J Chem Inf Model 56: 1399-1404.
- Daina A, Michielin O, Zoete V (2017) Swissadme: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7: 427-517.
- Lipinski C, Lombardo F, Dominy B, Feeney P (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46: 3-26.
- Ghose AK, Viswanadhan VN, Wendoloski JJ (1999) A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. A qualitative and quantitative characterization of known drug databases. J Comb Chem 1: 55-68.
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, et al. (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45: 2615-2623.
- Cortes C, Vapnik V (1995) Support vector machine. Mach Learn 20: 273-297.
- Molecular Operating Environment (MOE) (2008) Chemical Computing Group, Canada.
- Labute P (2009) Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins: Struct Funct Bioinf 75: 187-205.
- Goto J, Kataoka R, Muta H, Hirayama N (2008) Asedock-docking based on alpha spheres and excluded volumes. J Chem Inf Model 48: 583-590.
- Lipinski CA (2004) Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov Today Technol 1: 337-341.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 64: 4-17.
- Martin YC (2005) A bioavailability score. J Med Chem 48: 3164-3170.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences