Isolation and Identification of Multi Drug Resistant Efflux Pump Protein from Klebsiella pneumoniae
Raju Perla*
Department of Biotechnology, Godavari Institute of Engineering and Technology, Rajahmundry, Andhra Pradesh, India
- *Corresponding Author:
- Raju Perla
Godavari Institute of Engineering and Technology, Rajahmundry, Andhra Pradesh, India.
Tel: +919441469706
E-mail: Kingsolomon449@gmail.com
Received date: December 01, 2016; Accepted date: December 12, 2016; Published date: December 14, 2016
Citation: Perla R. Isolation and Identification of Multi Drug Resistant Efflux Pump Protein from Klebsiella pneumoniae. Pharm Biotechnol Curr Res. 2016, 1:1.
Abstract
Efflux pump in bacteria contribute to intrinsic resistance to a wide variety range of antibiotic and often have a broad substrate range due to the lack of new antibacterial agents. There is considerable interest in restoring the activities of older antibiotics by inhibiting the action of efflux pump and there being a usual area of active drug development by pharmaceutical companies, but the fact is that little is known about the interaction of such inhibitors with the bacterium. The addition of combinatorial drugs to the existing antibiotics can thereby enhance the activity and restore its effective use in treatment regimens. Keeping this in mind, the aim of current study was:
1. To isolate and identify a pathogenic multidrug resistant Klebsiella pneumoniae from clinical samples
2. To isolate and identify, the outer membrane multi drug resistant pump protein of isolated Klebsiella pneumoniae
Keywords
Efflux pump protein; Antibiotics; Hydrophobic drugs; Drug resistant bacteria; Multidrug transporters; Tripartite protein; Hydrophilic compounds
Introduction
Antibiotic resistance is the ability of a microorganism to withstand the effect of an antibiotic. Bacterial antibiotic resistances are an emerging and serious public health concern due to the compromised efficiency of antimicrobial agents used in the treatment of infectious diseases. If a bacterium carries several resistant genes, it is multi resistance.
Microorganism might exhibit resistance to drugs. The following are fairly well supported.
• Microorganism produced enzyme that destroy the active drug
• Microorganism change their permeability to the drug
• Microorganisms develop and altered the structural target for the drug
• Microorganisms develop and altered metabolic pathway that bypass the reaction inhibited by the drug
• Microorganisms develop an altered enzyme that can still perform its metabolic function. But is much less affected by the drug than the enzyme in the susceptible organism
• Efflux mechanism is to remove antibiotic
Efflux as a mechanism of antibiotic resistance was first reported linearly 1980’s for tetracycline [1,2]. Efflux pumps in bacteria contribute to intrinsic resistance to a wide range of antibiotics and often have a broad substrate range. Typically, the over expression of multidrug efflux pump confers resistance to antibiotic including fluoroquinolones, some dyes (e.g., Ethidiumbromide), detergents (e.g., SDS), and disinfectants (e.g., cetrimide). Genes encoding efflux pumps are typically chromosomally carried, ensuring the mutations are retained in further generations and will not be lost in the absence of selective pressure.
Antibiotic resistance to β-lactams, macrolides, quinolones and tetracycline is an increasing problem in many countries. The over expression of multi drug efflux pumps can lead to low level multidrug resistance, which possess a clinical problem. Multiple drug resistance (MDR) efflux pumps, which are coded by gene on the bacterial chromosome or on transmissible elements such as plasmids, are expressed in both Gram positive and Gram negative bacteria as this is attributed to the combined effect of the so called trance envelop efflux and reduced. The efflux pumps in Gram negative bacterium are more complex due to the presence of an outer membrane. They form a tripartite protein channel which requires a protein that traverses the periplasm known as the membrane fusion protein (MFP) and an outer membranelocated transporter.
Two major classes of multi-drug transporters
• ABC- type classes of multidrug transporters (utilise the free energy of ATP hydrolysis to pump drugs out of the cell).
• Secondary multidrug transporters (mediate the extrusion of structurally unrelated drugs in a coupled exchange with protons or sodium ions).
Secondary multidrug transporters
On the basis of size and similarities in the primary and secondary structures, the secondary multidrug transporters can be subdivided into four distinct families of transport proteins: The major facilitator super family (MFS) [3], The resistance nodulation cell division (RND) family [4,5]. The small multidrug resistance (SMR) family [6] and the toxic compound extrusion (MATE) family [7].
Major Facilitator Super family: The MFS consist of membrane transport proteins which are found in bacteria to higher eukaryotes and involved in the symport, antiport, or uniport of various substrates such as sugars, Krebs cycle intermediates, phosphate esters, oligosaccharides and antibiotics.
Small Multidrug resistance family: Multidrug transporters of the SMR family the smallest secondary efflux proteins known, are typically about 107 amino acid residues in length. Hydropathy analysis [6], gene fusions with alkaline phosphates and β-galactosidase. Alpha periodicity analysis, Cysteine accessibility scanning and fourier transform infrared spectroscopy support a model of tightly packed four helix antiparallel bundle. Due to the small size of the multidrug transporters of the SMR family, they function as homo oligomeric complexes. SMR family type proteins have been found in E. coli and B. subtilis.
Multidrug and toxic compound extrusion family: These proteins mediate resistance to dyes, hydrophilic compounds, fluoroquinolones and aminoglycosides. Norfloxacin accumulation experiments revealed that Vibrio parahaemolyticus contains an energy dependent efflux system designated as Nor M [8]. Nor M contains 12 putative TMS, and on this basis it has been suggested to be a member of the MFS family, but do not have any specific sequence similarity and do not exhibit any of the signature sequences.
Resistance- nodulation – Cell division family: Most lipophilic and amphilic drugs can cross the cytoplasmic membrane spontaneously and their accumulation on the periplasm would accelerate their re-entry into cytoplasm. To take full advantage of the exclusion properties of the cell wall in these bacteria and to by pass the periplasm it has been proposed that tripartite efflux systems consisting of a cytoplasmic membrane associated drugproton antiproton of the RND family, a periplasmic membrane fusion protein (MFB) and outer membrane channel forming protein are required to expel drug form the cytoplasm to the surrounding medium [9,10].
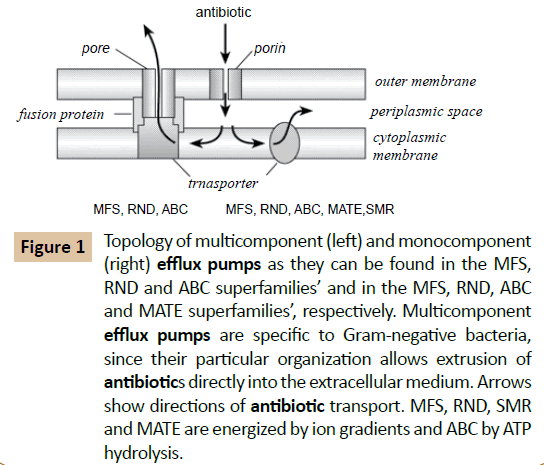
Although the exact molecular architecture of a tripartite RND efflux system still remains a mystery, some recent progress has been made regarding the topologies of the outer membrane protein component, Opr M and the RND transporter component Mex A, of one Pseudomonas Aeruginosa efflux pump, Mex ABOprM. Functional OprM is atrimeric protein besides the typical barrel configuration of the outer membrane embedded portions of the protein exhibits long periplasmic extensions capable of spanning the periplasm [11,12]. This led to the proposal that OprM is capable of directly interacting with the RND transporter Mex B. A topology analysis of Mex B using alkaline phosphatase fusions not only predicted 12 transmembrane domains typical of such transporter proteins but also several extensive periplasmic loops (Figure 1). Therefore it is conceivable that Opr M directly interacts with Mex B. The role of the MFP Mex AB plays in this process still remains a mystery, although it is clear that it is required for the functioning of RND efflux pumps [13].
Figure 1 Topology of multicomponent (left) and monocomponent (right) efflux pumps as they can be found in the MFS, RND and ABC superfamilies’ and in the MFS, RND, ABC and MATE superfamilies’, respectively. Multicomponent efflux pumps are specific to Gram-negative bacteria, since their particular organization allows extrusion of antibiotics directly into the extracellular medium. Arrows show directions of antibiotic transport. MFS, RND, SMR and MATE are energized by ion gradients and ABC by ATP hydrolysis.
Materials and Methods
Bacterial strain used
The cultures of pathogenic Klebsiella pneumoniae obtained from clinical laboratories were confirmed by performing standard microbiological and biochemical tests.
Confirmation of the bacterial strain using Gram staining
A drop of water was placed in centre of a clean grease free microscope slide. With an inoculating loop a small amount of bacterial culture was taken and smear was fixed. The slide was flooded with crystal violet staining reagent for 1 minute. The smear was then air dried and heat fixed. The slide was flooded with crystal violet staining reagent for 1 minute [5]. The smear was then washed in a gentle and direct steam of tap water for 2 seconds. The slide was flooded with iodine mordant for 1 minute and washed with 95% ethanol. The smear was then counter stained with saffranine for 30 seconds. The smear was again washed with a steam of tap water, air dried and examined under microscope.
Biochemical tests
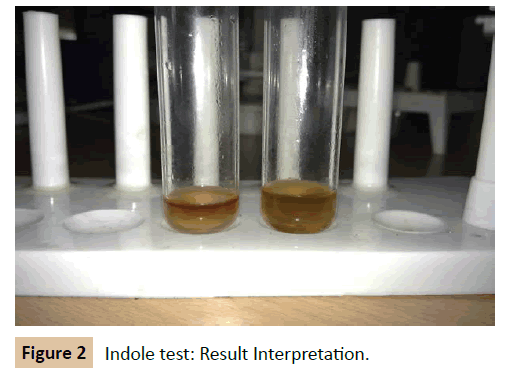
Indole test: A loopful of culture was taken and inoculated in the tubes of peptone broth. The tube was incubated at 37°C for 24-48 hrs. At the end of the incubation period 0.2 ml of Kovac’s reagent (for 5 ml of culture) was added and mixed thoroughly. The tubes were set aside for 5 minutes and observed for dark red violet colour in the top layer (Figure 2).
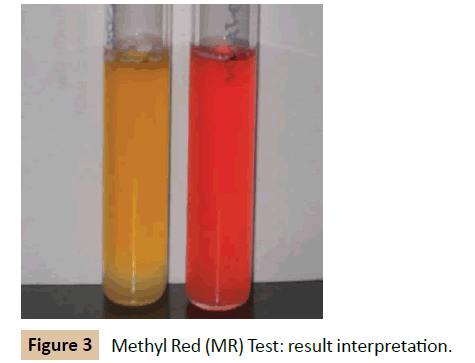
Methyl Red (MR) test: The culture was inoculated into glucose phosphate broth, which contains glucose and a phosphate buffer and incubated at 37°C for 48 hours. Over the 48 hours the mixedacid producing organism was observed for producing sufficient acid to overcome the phosphate buffer and remaining acid. The pH of the medium was tested by the addition of 5 drops of MR reagent. Development of red color is taken as positive. MR negative organism produces yellow color (Figure 3).
Voges Proskauer (VP) test: Bacterium was inoculated into glucose phosphate broth and incubated for at least 48 hours. 0.6 ml of alpha-naphthol was added to the test broth and shaken. 0.2 ml of 40% KOH was added to the broth and shaken. The tube was allowed to stand for 15 minutes. Appearance of red color is taken as a positive test. The negative tubes must be held for one hour, since maximum color development occurs within one hour after addition of reagents.
Citrate utilization test: Simmons citrate agar was prepared, sterilized at 121°C, for 15 minutes, and dispersed into test tubes, and slants were made. The test culture was streaked on the slant and incubated for 48 hours at 37°C and observed for growth.
A positive test shows a blue colour. Retention of original green colour and no growth on streak line indicates a negative growth.
Antibiotic sensitivity
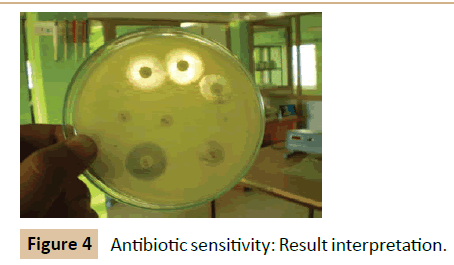
Kirby-Bauer method: Using an aseptic technique, a swab was moistened with broth culture of Klebsiella pneumoniae and spreaded uniformly over the surface of the nutrient agar plates by moving the swab back and forth in all directions (Figure 4). The antibiotic disc was removed form the container using sterile forceps and gently placed on the surface of the incubated plate. The disc was gently tapped with the forceps to fix it in position. The plates were incubated at 37°C for 24 hours right side up [14].
Determination of MIC: Minimum inhibitory concentration (MIC) was determined by serial two fold dilution in LB Broth by using the broth dilution method. Results were determined after incubation at 37°C for 18 hours. 100 μl aliquots of log-phase cells grown in LB broth were added to an equal volume of LB broth containing serial two-fold dilution of antibiotic. Following incubation at 37°C for 18 hrs, growth was assessed visually and to MIC was reported as the lowest concentration of antibiotic inhibiting visible growth.
OMP preparation: OMP were prepared by modified method of Porton. The strain K. pneumoniae were grown in LB broth minimal inhibitory concentrations of antibiotic such as rifampicin (10 μg), and ampicillin (5 μg) and incubated at 37°C for 12 hrs. The cells were pelletized and washed twice with sterile PBS and were suspended in 10 mM tris-Hcl (pH 7.8), 5 mM EDTA and 1 mM 2-mercaptoethanol. The cells were broken by mechanical lysis and membranes were subsequently pelleted out by centrifugation at 5000 rpm for 10 minutes. The membrane pellet was resuspended in a solution containing 1% Triton-X 100 and 1mM 2-mercaptoethanol and incubated overnight at 4°C. Insoluble membrane proteins were pelleted by high speed centrifugation at 100000 g for 1 hour and resuspended in distilled water.
Protein estimation: The amount of protein was estimated by Lowry’s method. The blue colour developed by reaction between Folin Ciocalteau reagent and protein samples were read at OD 660 nm (ELICOSL177). A set of Bovine Serum Albumin standard was also performed [15]. The concentration of protein sample was calculated from the standard graph and loaded on SDS polyacrylamide gel electrophoresis in Figure 5 (SDS PAGE).
SDS-PAGE: Principle: Anionic detergent, SDS in combination with a reducing agent and heat dissociates the proteins. SDSPolypeptide complexes migrate through polyacrylamide gels in accordance with the size of the polypeptide when subjected to an electric field.
Procedure: Glass plates used were cleaned and where assembled with appropriate spacers 5 ml of separating gel was poured and covered with a layer of distilled water. After polymerization the distilled water was removed and 2 ml of stacking gel was poured and a comb was inserted. After 10-15 minutes the comb was carefully removed and the wells were washed with water. This was transferred to the electrophoretic apparatus. 10 μl of gel loading buffer was added to the 20 μl of sample protein. The mixture was heated at 100°C for 7 minutes in heating block. The sample was loaded in the well. The gel was run at 100 V for 1.5 hours. After the run, stacking gel was removed and stained in Coomassie Brilliant Blue for 4 hours and followed by destaining using a mixture of methanol, glacial acetic acid and water.
The molecular weight of the over expressed protein was determined by comparing the relative molecular mass along with standard marker proteins Bovine serum albumin (66 KDa), Ovalbumin (43 KDa), Carbonic anhydrase (29 KDa), Lysozyme (14.3 KDa).
Results
Microscopic examination and biochemical reactions of Klebsiella pneumonia
The culture of Klebsiella pneumoniae obtained from clinical laboratories was confirmed with the following result of microscopic examination and biochemical tests.
Gram staining: It was observed as Gram negative rod shaped bacterium.
Biochemical reactions of K. pneumonia: The cultures of K. pneumoniae obtained from clinical laboratories were confirmed with the following result of biochemical tests. Four biochemical tests were employed. Indole, Methyl red, Voges Proskauer and Citrate utilization test. The organism gave --++ test result. It means for negative for both indole and Methyl red test and positive result for Voges Proskauer test and Citrate test.
Antibiotic sensitivity test
The organism was found to be resistant against four antibiotic and sensitive for four. The pattern is given in the Table 2.
| S. No. | Antibiotic | Amount per disc | Diameter of the zone of inhibition | ||
|---|---|---|---|---|---|
| Resistance (mm or less) | Intermediate (mm) | Sensitive (mm or more) | |||
| 1 | Ampicillin | 10 mcg | 3 | 14 -16 | 17 |
| 2 | Ciprofloxacin | 10 mcg | 15 | 16-20 | 21 |
| 3 | Chlororamphenical | 10 mcg | 12 | 13-17 | 18 |
| 4 | Erythromycin | 10 mcg | 13 | 14-22 | 23 |
| 5 | Penicillin | 10 mcg | 19 | 22-27 | 28 |
| 6 | Rifampicin | 15 mcg | 16 | 17-18 | 19 |
| 7 | Streptomycin | 10 mcg | 11 | 12-14 | 15 |
Table 1 Standard table for zones of inhibition (Himedia).
| Ampicillin | Resistant |
|---|---|
| Streptomycin | Sensitive |
| Chloramphenicol | Sensitive |
| Ciprofloxacin | Sensitive |
| Rifampicin | Resistant |
| Erythromycin | Sensitive |
| Penicillin | Resistant |
Table 2 The pattern of Antibiotic sensitivity test.
Estimation of protein
By employing Lowry‘s method, the total protein concentration of isolated outer membrane proteins was found to be 200 μg/ml.
SDS
The molecular weight of the over expressed protein was calculated by comparing with protein marker and control. A 66 KDa protein was found to be over expressed in the lane corresponding to those of MDR Klebsiella pneumoniae with antibiotics, when compared to those without antibiotic.
Discussion
The evolution of drug resistance among pathogenic strains have rendered many treatment regimens inactive and posed a big threat to clinicians across the globe. The major mechanism behind the emergence of drug resistance has attained considerable attention among researchers.
Efflux mediated drug resistance in microorganism gains prior significance via the unique property of efflux protein to pump out antibiotics of same or different families to a greater intent has shown the significance of drug resistant pumps [16]. One of the aim of drug resistance study was to evaluate the presence of outer membrane drug resistant proteins in clinical. The samples are confirmed as Klebsiella pneumoniae by performing biochemical and staining techniques. The organism was found to be gram negative rod shaped and it gave (++--) result for IMViC test [17].
The isolation and characterisation of drug resistant pumps gives opportunity for designing inhibitors which can inhibit the membrane pumps, thereby bring back antibiotics of old generation effectively.
The intensive research on novel inhibitors against multidrug efflux pump had led to the discovery of many natural inhibitors such as reserpine, piperine and berberine. But little is known about the molecular mechanism under lying in the same.
The efflux reviewed to be one major mechanism of drug resistance in Klebsiella pneumoniae and any inhibition in Nor A pumps can make the organism sensitive to common antibiotics (Table 1). The current study put forward many suggestions to combat drug resistance mediated by efflux pumps. The further molecular characterization of the protein by sequencing bioinformatics tools can give opportunity to design ligand (chemical antagonists) by docking and theoretical calculations.
Conclusion
Multi drug resistance exhibited by pathogenic strains of bacteria remain always a threat for existing treatment regimens. A pathogenic bacterial strain Klebsiella pneumoniae exhibiting multidrug resistance via expressing outer membrane protein efflux pumps was isolated and identified having an over expressed outer membrane efflux pump protein of approximately molecular weight of 66 KDa. The structure of this protein can be determined using bioinformatics tools and an effective drug can be designed to get rid of multi drug resistant bacteria.
References
- Ball Y, Lemieux MJ, Song, J, Auer M, Wang DN (2003) Structure and mechanism of glycerol 3 phosphate transporter from Escherichia coli. Science (Wash DC) 301: 616-620.
- McMurry L, Petruai RE, Levy SB (1980) Active efflux of tetracycline encoded by four genetically different tetracycline resistant determinants in Escherichia coli. Proc. Nat. Acad. Sci. USA 77: 3974-3977.
- Pao SS, Paulsen IT, Sair MH (1998) Major facilitator super family. Microbiol. Mol. Biol. Rev. 62: 1-34.
- Saier MH (2000) An functional phylogenetic classification system for trans membrane solute transporters. Microbiol Mol Biol Rev 64: 354-411.
- Nikaido H (1998) Antibiotic resistance caused by Gram-negative antibiotic efflux pumps. Clin Infect Dis 27: 532-541.
- Paulsen IT, Brown MH, Skurray RA (1996) Proton dependent multi drug efflux systems. Microbiol Rev 60: 575-608.
- Brown MH, Paulsen IT, Skurray RA (1999) The multidrug efflux protein Nor M is a prototype of a new family of transporters. Mol Microbiol 31: 394-395.
- Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, et al. (1998) Nor M, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42: 1778-1782.
- Nishino K, Yamaguchi A (2001) Over expression of the response regulator. EvgA of the two component signal transduction system modulates multi drug resistance conferred by multidrug resistance transporters. J Bacteriol 183: 1455-1458.
- Martinez JL, Baquero B (2002) Interactions among stratergies associated with bacterial infection: Pathogenicity, epidemiology and antibiotic resistance. lin Microbiol Rev 15: 647-649.
- Huda MN, Chen J, Morita Y, Kuroda T, Mizushima T, et al. (2003) Gene cloning and characterization of VcrM, a Na+ - coupled multidrug efflux pump, from Vibrio cholerae non-O1. Microbiol Immunol 47: 419-427.
- Jack DL, Storms ML, Tchieu JH, Paulsen IT, Saier MH (2000) A broad specificity multidrug efflux pumps requiring a pair of homologous SMR type of proteins. J Bacteriol 182: 2311-2313.
- George AM, Levy SB (1983) Amplifiable resistance to tetracycline, Chloramphenicol and other antibiotics in Escherichia coli. Involvement of a non-plasmid determinant efflux of tetracycline. J Bacteriol 155: 531-540.
- Barbosa T, Levy SB (2000) The impact of antibiotic use on resistance development and persistence. Drug Resistant. Update (Medline) 3: 303-311.
- Paulsen IT, Skurray RA (1993) Topology, structure and evolution of two families of proteins involved in antibiotic and antiseptic resistance in eukaryotes and prokaryotes - an analysis. Gene 124: 1-11.
- Chung YJ, Saier MH (2001) SMR type multi drug resistant pumps. Curr Opin Drug Disc Dev 4: 237-245.
- Van VHW, Konings WN (1998) The ABC family of multi drug transpoters in microorganisms. Biochem Biophys Acta 1365: 31-36.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences