ISSN : 2249 - 7412
Asian Journal of Plant Science & Research
Investigating Genetic Diversity of Kabuli Chickpea (Cicer arietinum L.) Genotypes in Awabel District, Northwestern Ethiopia
Gebremeskel Mequanint Mulu* and Ahadu Menzir Anley
Department of Plant Science, Debre Merkos University, Debre Markos, Ethiopia
- *Corresponding Author:
- Gebremeskel Mequanint Mulu
Department of Plant Science,
Debre Merkos University,
Debre markos,
Ethiopia,
Tel: 251931548104;
E-mail: gebremequanint26@gmail.com
Received date: June 14, 2023, Manuscript No. AJPSKY-23-16974; Editor assigned date: June 19, 2023, PreQC No. AJPSKY-23-16974 (PQ); Reviewed date: July 04, 2023, QC No. AJPSKY-23-16974; Revised date: September 14, 2023, Manuscript No. AJPSKY-23-16974 (R); Published date: September 22, 2023, DOI: 10.36648/2249-7412.13.9.313
Citation: Mulu GM, Anley AM (2023) Investigating Genetic Diversity of Kabuli Chickpea (Cicer arietinum L.) Genotypes in Awabel District, Northwestern Ethiopia. Asian J Plant Sci Res Vol:13 No.9:313
Abstract
The objective of the study was to assess the magnitude of genetic diversity of 36 Kabuli Chickpea genotypes including two varieties in 6 × 6 simple lattice design in 2020/2021 main cropping season at Awabel district. 36 genotypes were grouped in to seven (7) distinct clusters based on the 13 (thirteen) morphological traits. The maximum intra-cluster distance was observed in cluster-IV, indicates the genotypes included in this cluster were more divergent than other clusters. The maximum inter cluster distance were observed between cluster I and II, followed by cluster I and III, between cluster-I and VII and between cluster I and IV. The genotypes which had large inter cluster distance may give high heterotic response resulting in better recombinants. The first six principal components explained about 87.22 % of the total variation. Seed yield, number of seeds per plant and number pods per plant was major contributors of variation for (PC1). Overall, the present study indicates the presence of wide range of diversity among the tested genotypes it is vital role for further use in breeding program.
Keywords
Chickpea; Cluster; D2 statistics; Genotypes; Principal component
Introduction
Chickpea (Cicer arietinum L.) is one of the most important annual food grain legumes and first domesticated grain legume cultivated in more than 57 countries under varied environmental conditions throughout the world [1]. Chickpea is one of the major pulse crop grown in Ethiopia under wide ranging agro ecological conditions [2]. The crop is considered as highly nutritious and an inexpensive source of proteins, carbohydrates, fats and is a good source of minerals and essential with enhanced levels of essential dietary nutrients for many people and generating income for small households [3].
However, the national average seed yield in Ethiopia is low, 2.18 tonsha-1 far below its genetic potential yield of while under best adoption it gave 3.5 tons ha-1, because the production of the crop nationally constrained by usage of in appropriate improved varieties, use of inherent low productive farmers’ varieties, biotic and a biotic factors [4]. Better and more stable yields are the major goals of plant breeding programs. Such enhancement of crops needs conception and introduction of genetic diversity, inbreeding coupled with selection and extensive evaluation of breeding resources at multiple locations over years to identify adapted and stable genotypes with desirable traits [5]. The inception of chickpea breeding program has boarded on understanding the challenges of chickpea production and looking for genetic improvement basis to curb some of those key gaps. Information regarding genetic diversity is vital to determine breeding materials for further breeding programs. Diversity among the genetic resources is the base for any genetic improvement in crop plants. The availability of genetic variability in a gene pool is a prerequisite for a successful breeding program to achieve the expected genetic improvement through selection. Estimating genetic diversity of Kabuli chickpea accessions help in the determination of highly diversified germplasm which provides ample opportunity to breeders to look for desirable traits for developing new and super performing varieties. Evaluation of genetic diversity among genotypes is the breeder’s duty [6].
Principal Component Analysis (PCA) reflects the importance of the largest contributor to the total variation at each axis for differentiation [7]. PCA was used to find out the traits, which accounted more for the total variation. Genetic diversity studies had been conducted by different researchers and prove the presence of high genetic variation among studied genotypes. Despite, Ethiopia is well thought out the secondary center of diversity for chickpea; genotypic diversity is worthless it is promptly conserved and efficiently utilized in crop improvement programme. An evidence behind is up to date about 29 super performing varieties were released in both types since 1970’s. Yet, most of the developed verities were also Desi type. Previous information’s available on genetic variation is limited and there is lack of availability of genetic diversity to improved Kabuli chickpea varieties [8].
This indicates conducting such an original research specific to a site will have an important role on generating genetic information on diversity of genotypes based on morpho agronomic traits for further improvement through selection and/or hybridization. The aim of the study was:
• To assess the level of genetic diversity among examined Kabuli chickpea genotypes
• To determine the important traits contributing more to the total variation.
Materials and Methods
Description of experimental site
The genotypes were evaluated at experiment site of Awabel district, East Gojjam Zone, during 2021 main cropping season. The site laid 10°29’ latitude N and 37°44’ longitude with an altitude of 2104 m.a.s.l. The average mean annual rainfall was 1090 mm and its minimum and maximum temperatures are 15°C and 24°C. The soil was black vertisol with pH value 6.45 (ADAO).
Planting material
A total of 36 Kabuli chickpea genotypes including two standard check varieties (Arerti and Habru) were included in the study.
The experimental materials were obtained from the Highland Pulse Research Program, Debre Zeit Agricultural Research Center (DZARC) (Table 1).
| Code | Genotype pedigree | Code | Genotype pedigree | Code | Genotype pedigree |
|---|---|---|---|---|---|
| G-1 | FLIP-93-93C | G-13 | Arerti | G-25 | FLIP-12-343C |
| G-2 | FLIP-12-53C | G-14 | FLIP-12-60C | G-26 | FLIP-12-311C |
| G-3 | FLIP-12-110C | G-15 | FLIP-12-57C | G-27 | FLIP-12-176C |
| G-4 | FLIP-12-37C | G-16 | FLIP-12-86C | G-28 | FLIP-12-40C |
| G-5 | Habru | G-17 | FLIP-12-342C | G-29 | FLIP-12-01C |
| G-6 | FLIP-12-198C | G-18 | FLIP-12-263C | G-30 | FLIP-12-331C |
| G-7 | FLIP-12-107C | G-19 | FLIP-82-150C | G-31 | FLIP-12-265C |
| G-8 | FLIP-12-287C | G-20 | FLIP-88-85C | G-32 | FLIP-12-55C |
| G-9 | FLIP-12-06C | G-21 | FLIP-12-108C | G-33 | FLIP-12-197C |
| G-10 | FLIP-12-18C | G-22 | FLIP-12-322C | G-34 | FLIP-12-210C |
| G-11 | FLIP-12-79C | G-23 | FLIP-12-310C | G-35 | FLIP-12-75C |
| G-12 | FLIP-12-61C | G-24 | FLIP-12-109C | G-36 | FLIP-12-192 |
Table 1: List of experimental materials used.
Experimental design, procedures and trial management
The experiment was carried out in 6 × 6 simple lattice designs. The distance between replications, blocks and plots were 2 m, 1 m and 0.5 m, respectively. The total net plot size of were 1.35 m2. Each genotype was planted with spacing of 30 cm between rows and 10 cm between plants.All agronomic practices were done as per the recommendation for chickpea production.
Data collected
The data were collected from the net plot area within three middle rows at randomly selected and tagged 5 individual plants by adopting [9]. Thirteen morph agronomic trait data were collected. These are Days to 50% Flowering (DF), Days to 90% Maturity (DM), Pod Filling Period (PFP), Hundred Seeds Weight (HSW) in gm, Biomass Yield (BY) in Kgha-1, Seed Yield (SY) in kgha-1 and Harvest index (HI), Plant Height (PH) in cm, Number of Primary Branch (NPB), Number of Secondary Branches (NSB), Number of Pods Plant-1 (NPP) and Number of Seeds Per Pod (NSP).
Data analysis
Genetic divergence analysis: Genetic divergence analysis was computed based on multivariate analysis using Mahalan obis’s D2 statistic by SAS Software program 9.4.
Estimation of genetic distance: Genotypes are clustered based on similarity of characters based on the squared distances (D2) values using statistical technique developed by Mahalanobis, et al. used to classify the different genotypes into different groups. Clustering of genotypes was done using Tocher’s method as described by Singh and Chaudhary, et al. The Squared distances (D2) for each pair of genotype combinations was computed using the following formula:
D2ij=(Xi- Xj) S-1 (Xi – Xj)
Where,
D2ij= The square distance between any two genotypes i and j, Xi and Xj= The vectors for the values for genotype ith and jth genotypes, and S-1= the inverse of pooled variance covariance matrix.
Estimation of intra and inter-cluster distances: Average intra and inter cluster D2 values was estimated using the formula where ΣDi2 is the sum of distance between all possible combinations (n) of the genotypes included in a cluster. Significance of the squared distances for each cluster was tested against the tabulated χ2 values at p degree of freedom at 5% probability level.
Where,
p=number of characters used for clustering genotypes.
• Average intra and inter cluster D2 values
• Average intra cluster D2, D2= Σ Di2/n
Where,
ΣDi2 is sum of distances between all possible combinations (n) is the population included in a cluster.
Average inter cluster D2, D2=ΣDi2/ni.nj
Where,
ni=number of population in cluster
i, nj=number of population in cluster j
Principal Component Analysis (PCA): The principal component analysis was done to recognize the characters contributing more to the total variation using correlation carried by SAS jmp statistical software (2016). Principal components with eigenvalues greater than one are considered in this analysis according to Chahal and Gosal, et al. Principal components were computed using SAS computer software based on formulas suggested by Holland, et al. The first PCA value (Y1) is given by the linear combination of the variables X1, X2 ...Xp Y1=a11X1 +a12X2+...+a1pXp
The next principal component is computed in the same way, Y2=a21X1 +a22X2 +...+a2pXp this continues until a total of p principal components have been computed.
Results and Discussion
Genetic divergence analysis
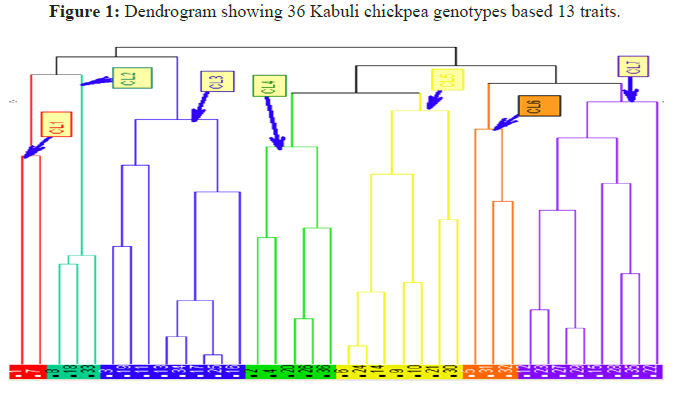
The distribution of 36 Kabuli chickpea genotypes classified into seven distinct clusters based on similarity and difference of traits (Table 2 and Figure 1).
| No of cluster | Name of genotypes Pedigree | No of genotypes | Percent |
|---|---|---|---|
| I | FLIP-93-93C and FLIP-12-107C | 2 | 5.56 |
| II | FLIP-12-287C, FLIP-12-263C and FLIP-12-197C | 3 | 8.33 |
| III | FLIP-12-110C, Arerti, FLIP-12-86C, FLIP-12-342C, FLIP-82-150C, FLIP-12-79C, FLIP-12-343C and FLIP-12-210C | 8 | 22.22 |
| IV | FLIP-12-53C, FLIP-12-37C, FLIP-88-85C, FLIP-12-311C and FLIP-12-192 | 5 | 13.89 |
| V | FLIP-12-198C, FLIP-12-06C, FLIP-12-18C, FLIP-12-60C, FLIP-12-109C, FLIP-12-108C and FLIP-12-331C | 7 | 19.44 |
| VI | Habru, FLIP-12-265C and FLIP-12-55C | 3 | 8.33 |
| VII | FLIP-12-75C, FLIP-12-61C, FLIP-12-176C, FLIP-12-40C, FLIP-12-01C, FLIP-12-322C, FLIP-12-310C and FLIP-12-57C | 8 | 22.22 |
Table 2: Distribution of 36 Kabuli Chickpea genotypes in seven different clusters based on square distance.
Clustering analysis of genotypes: Cluster III and VII each containing 8 genotypes and cumulatively accounts 44.44% of the total genotypes evaluated. Cluster-V included 7 genotypes and covers 19.44% of total genotypes tested, cluster- IV contained 5 genotypes (13.89%), cluster-II and VI each contained 3 genotypes with 8.33 % share and cluster-I consists of a minimum number of genotypes 2 (5.56%) genotypes. The result indicates that the presence of wide range of diversity among the tested genotypes.
Cluster distance analysis: The intra and inter cluster distance showed the existence of significant genotypic difference among the clusters and within clusters except cluster VI and II showed non-significant intra cluster distance as tested by chi-square distribution (Table 3). The maximum intra cluster distance was recorded in cluster- IV (D2=82.98) followed by cluster III (D2=80.74) and the lowest for cluster-VI (D2=17.22). The highest intra cluster distance of cluster- IV, indicates the genotypes included in this cluster were more divergent than other clusters. The maximum inter-cluster distance were observed between cluster I and II (D2=208791720) followed by cluster I and III (D2=172892422), between cluster-I and VII (D2=123754371) and between cluster I and IV (D2=101947039), this indicates the presence of maximum genetic difference among the tested genotypes. Whereas, the lowest inter cluster distance was observed between cluster IV and V (D2=4262487), indicates existence of closer proximity between these clusters as comparatively with other clusters.
| Clusters | I | II | III | IV | V | VI | VII |
|---|---|---|---|---|---|---|---|
| I | 78.2 | 208791720 | 172892422 | 101947039 | 68543967 | 45608626 | 123754371 |
| II | 23.64 | 655 | 20758478 | 38077469 | 64537215 | 36351036 | |
| III | 80.74 | 71691491 | 60827411 | 51940169 | 9296483 | ||
| IV | 82.98 | 4262487 | 19224177 | 29735071 | |||
| V | 68.58 | 5482395 | 22905005 | ||||
| VI | 17.22 | 21162690 | |||||
| VII | 47.88 |
Note: X2= 26.22 at 5% probability level
Table 3: Intra (diagonal) and inter (off diagonal) square distance (D2) values of 36 chickpea genotypes.
Cluster mean analysis: The mean value of the 13 traits in each cluster is presented in Table 4. Among cluster means Cluster (I) was characterized by maximum biomass yield (9188.19 kgha-1), highest seed yield (3273.19 kgha-1), more number of pod per plant (40.32), more number of seed per pod (1.27), highest number of primary branch (5.23), number of seed per plant (45.73), number of secondary branch (9.0) and the longest plant height (51.99 cm). Genotypes in this cluster showed the best performance for desirable agronomic traits above ground biomass yield, number of pod per plant, number of seed per pod, number of primary branch, number of seed per plant and number of secondary branch. Cluster-V characterized intermediate mean value for all traits.
| Traits | CL1 | CL2 | CL3 | CL4 | CL5 | CL6 | CL7 |
|---|---|---|---|---|---|---|---|
| DF | 55.34 | 57.84 | 56.68 | 52.99 | 55.74 | 56.13 | 56.36 |
| PFP | 71.53 | 66 | 63.42 | 67.26 | 67.23 | 65.88 | 67.57 |
| DM | 127.3 | 123.21 | 119.83 | 120.9 | 123.01 | 122.13 | 123.83 |
| PH | 51.99 | 48.47 | 44.96 | 47.33 | 46.66 | 42.55 | 51.69 |
| NPB | 5.23 | 2.97 | 4.49 | 4.27 | 3.56 | 3.13 | 3.23 |
| NSB | 9.01 | 7.18 | 7.29 | 8.42 | 6.81 | 6.69 | 6.39 |
| NPP | 40.32 | 33.21 | 35.33 | 34.8 | 33.36 | 27.64 | 32.29 |
| NSP | 1.27 | 1.15 | 1.2 | 1.12 | 1.16 | 1.09 | 1.23 |
| NSPP | 45.73 | 31.9 | 43.55 | 36.47 | 34.69 | 39.53 | 40.92 |
| BY | 9188.19 | 7946.41 | 6926.92 | 8642.31 | 7005.15 | 7356.63 | 6517.27 |
| HSW | 27.16 | 29.11 | 29.28 | 37.22 | 34.29 | 29.59 | 33.15 |
| SY | 3273.19 | 2837.61 | 3013.2 | 2942.09 | 2829.14 | 2196.95 | 2610.65 |
| HI | 0.355 | 0.38 | 0.46 | 0.41 | 0.414 | 0.34 | 0.435 |
Note: DF: Days to Flowering; PFP: Pod Filling Period; DM: Days to Maturity; PH (cm): Plant Height in cm; NPB: Number of Primary Branches; NSB: Number of Secondary Branches; NPP: Number of Pods Per Plant; NSP: Number of Seeds Per Pod; NSPP: Number of Seeds Per Plant; BY: Biomass Yield in kgha-1; HSW (g): Hundred Seed Weight in gram; SY: Seed Yield in kgha-1; HI=Harvest Index.
Table 4: Mean values of 13 traits of the seven clusters of 36 Kabuli chickpea genotypes.
Principal component analysis
The six Principal Components (PC) from PC1 to PC6 with eigenvalues greater than one contributed about 87.22% of the total variation, with PC1, PC2, PC3, PC4 PC5 and PC6 in that order explaining 24.1%, 18.86%, 15.57%, 10.97%, 9.37% and 8.36% of the gross variation among 36 Kabuli chickpea genotypes evaluated for 13 traits. The major contributors of variation for (PC1) were seed yield, number of seeds per plant and number pods per plant. Number of primary branches, number of secondary branches and harvest index explain the highest variation on PC2. Highest contributors for explained variance in PCA3 include days to flowering, pod filling period and days to maturity (0.61). Variation in PC4 was mainly due to hundred seed weight (0.56); pod filling period (0.36) and number of secondary branches (0.33). Plant height (0.53), number of primary branches (0.36) and secondary branches (0.299) contributed more in PCA5. Plant height (0.56), harvest index (0.54) and number seeds per pod (0.31) explain the highest variation on PC6.
Discussion
Tested genotypes were genetically divergent based on divergence analysis
In present study, we were evaluating the magnitude of genetic diversity among 36 Kabuli Chickpea genotypes for 13 agro morphology traits. Hence, the main discussions for the studied traits are described below. Kabuli chickpea genetic resources represent an invaluable source of genetic diversity that is predictable to be highly useful for existing and future breeding program [10,11]. Achievements in genetic improvement of any crop is greatly depends on the availability of genetic resources and the extent of genetic diversity among important traits. Information of genetic diversity among genotypes would have contribution to chickpea breeders to develop the most adaptive, better and more productive varieties through hybridization.
Seven distinct clusters showed wide range of diversity: From the result in Table 3, 36 Kabuli Chickpea genotypes were grouped into seven different clusters based on square distance, indicating that the presence of wide range of diversity among the tested genotypes.
Distance analysis revealed the highest intra and inter cluster distances: The results illustrated in the above revealed the highest intra cluster distance of cluster-IV, indicates the genotypes included in this cluster were more divergent than other clusters. The genotypes which had large inter cluster distance between cluster I and II (D2=208791720) followed by cluster I and III (D2=172892422) can be used for hybridization program and may give high heterotic response resulting in better recombinants. Whereas, genotypes which had the lowest inter cluster distance was, indicating existence of closer proximity between them. Maximum genetic recombination and variation in the subsequent generation is predicted from crosses that involve parents from the clusters characterized by maximum distances, crosses between genotypes of cluster I with II, cluster I with III are expected to be the best scenario to develop better genetic recombination and generate desirable segregants with broad genetic base. Hence, selection in segregating generations of these crosses seems to give promising results. Similar findings were noted by Aarif, et al.; Temesgen, et al.; Mayuriben, et al.; Thakur, et al. in chickpea genetic diversity study. Temesgen, et al. revealed grouping 49 Kabuli chickpea genotypes in to 8 clusters and indicted the diversity of their studied genotypes [12].
Cluster mean analysis showed the performance of desirable traits in each cluster: The mean value of the 13 traits in each cluster is presented in Table 4. The cluster mean value of genotypes in cluster (I) showed the best performance for desirable agronomic traits above ground biomass yield, number of pod per plant, number of seed per pod, number of primary branch, number of seed per plant and number of secondary branch. This might be the best situation for Kabuli chickpea improvement through selection and hybridization of desired traits. As nake and Dagnachew, et al. revealed first level priority as far producers put productivity, resistance, adaptation traits and to be followed by market and taste [13-16]. In line with these from cluster mean values, genotypes in cluster I deserve consideration for their direct use as parents in hybridization programs to develop high yielding varieties. The current finding exerted the average cluster mean of 13 characters revealed that none of the clusters contained genotype with all the desirable characters and so recombinant breeding between genotypes of different clusters are needed. In addition, to create genetic diversity in diverse desirable traits, multiple crosses are performing using these divergent parental lines may give good results. Similarly, Chahal and Gosal, et al., Temesgen, et al., Sintayehu and Gize, et al., Thakur, et al. and Asnake and Dagnachew, et al. stated that selection of parents should also considerthe superior advantage of each cluster and each genotype within a cluster depending on specific objective of hybridization [17,18].
Principal component analysis explained total variations: The consequences of the principal components analysis in Table 5 showed that the first six principal components with eigenvalue greater than one elucidated 87.22% of the total variation among genotypes [19]. The major positive contributors of variation for (PC1) were seed yield, number of seeds per plant and number pods per plant as contributors. The first principal component accounted more (24.1%) for the total variation in the figures compared to succeeding components. The second principal component was positively associated with number of primary branches, number of secondary branches and harvest index. The results designated that latent candidate traits for breeding materials could be obtained from genotypes in the first two principal components PC1 and PC2 contributed more to the total variation and it suggested that many opportunities for genetic improvement through selection than the least succeeding principal components. In line with the present finding, Temesgen, et al.; Fasil and Zerafu, et al. revealed similar results [20]. Overall, the results from this finding wide genetic diversity among the tested genotypes important for successful breeding program with respect to most studied traits [21-24].
| Traits | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 |
|---|---|---|---|---|---|---|
| DF | -0.21 | 0.22 | 0.28 | -0.56 | 0.18 | -0.047 |
| PFP | 0.013 | 0.15 | 0.54 | 0.36 | -0.32 | 0.0032 |
| DM | -0.11 | 0.26 | 0.61 | -0.051 | -0.15 | -0.03 |
| PH | -0.005 | -0.095 | 0.26 | -0.021 | 0.53 | 0.56 |
| NPB | 0.28 | 0.43 | -0.13 | 0.16 | 0.36 | 0.049 |
| NSB | 0.19 | 0.43 | -0.083 | 0.33 | 0.299 | -0.23 |
| NPP | 0.37 | 0.32 | 0.13 | -0.099 | 0.011 | -0.21 |
| NSP | 0.38 | -0.13 | 0.13 | -0.2 | 0.18 | 0.31 |
| NSPP | 0.42 | 0.018 | -0.11 | 0.018 | -0.36 | 0.24 |
| BY | 0.37 | -0.35 | 0.17 | -0.1 | 0.12 | -0.31 |
| HSW | -0.016 | -0.28 | 0.196 | 0.56 | 0.1 | 0.19 |
| SY | 0.47 | -0.15 | 0.11 | -0.19 | -0.22 | 0.037 |
| HI | -0.035 | 0.37 | -0.19 | -0.077 | -0.33 | 0.54 |
| Eigenvalue | 3.13 | 2.45 | 2.02 | 1.42 | 1.22 | 1.09 |
| Percent | 24.1 | 18.86 | 15.57 | 10.96 | 9.37 | 8.36 |
| Cum Percent | 24.1 | 42.96 | 58.53 | 69.49 | 78.86 | 87.22 |
Note: PC: Principal Component; DF: Days to Flowering; DM: Days to Maturity; PFP: Pod Filling Period; PH: Plant Height; NP: Number of Primary Branches; NSB: Number of Secondary Branches; NPP: Number of Pod Per Plant; NSPP: Number of Seed Per Plant; NS: Number of Seed Per Pod; BY: Biomass Yield; HSW: Hundred Seed Weight; SY: Seed Yield and HI: Harvest Index.
Table 5: Eigenvectors, eigenvalues and percentage of total variance explained by the first six Principal Components (PC) for 13 traits in 36 chickpea genotypes.
Conclusion
In conclusion, wide genetic diversity were observed among genotypes between clusters, indicates crossing of genotypes in these cluster might be the best situation to develop moderately better genetic recombination and generate desirable segregants. The first six Principal Components (PC) explained about 87.22% of the total variation among Kabuli chickpea genotypes. The present result it has been observed adequate existence of diversity for most of the traits in the studied genotypes which need to be exploited in future Kabuli chickpea breeding. However, this study was conducted for one season and at one location which needs to be conducted in subsequent breeding trials considering more locations to develop high yielding varieties and marketable produce for progress improvement for further Kabuli chickpea breeding.
Data Availability
The data used to support the findings of this study are included in the supplementary information file.
Conflicts of Interest
No conflicts of interest.
Acknowledgements
The authors would like to express their great appreciation to the Ministry of Education, Debre Markos University; Highland Pulse Research Program, DZARC and Awabel District Agricultural Office, for facilitating the research process.
References
- Aarif M, Rastogi NK, Johnson PL, Yadav SK (2017) Genetic divergence analysis in kabuli chickpea (Cicer arietinum L.). J Pharmacogn Phytochem 6:1775-1778
- Aditya JP, Bhartiya P, Bhartiya A (2011) Genetic variability, heritability and character association for yield and component characters in soybean (G. max (L.) Merrill). J Cent Eur Agric 12:27-34
- Ali Q, Ahsan M, Farooq J (2010) Genetic variability and trait association in chickpea (Cicer arietinum L.) genotypes at seedling stage. Electron J Plant Breed 1:334-341
- Ali Q, Tahir MH, Sadaqat HA, Arshad S, Farooq J, et al. (2018) Genetic variability and correlation analysis for quantitative traits in chickpea genotypes (Cicer arietinum L.). Int Sch J 12:1–4
- Tsehaye A, Fikre A (2020) Genetic diversity analysis for some desi type chickpea (Cicer arietinum L.) advanced lines under potential environment of North Gondar, Ethiopia. Am J Biosci Bioieng 8:27-35
- Fikre A, Bekele D (2019) Chickpea breeding and crop improvement in Ethiopia: Past, present and the future. Univers J Agric Res 8:33-40
- Asnake Fikre (2014) An overview of chickpea improvement research program in Ethiopia. J Int Leg Soc 3:34-41
- Merga B, Haji J (2019) Economic importance of chickpea: Production, value and world trade. Cogent food agric 5:1615718
- Choudhary P, Khanna SM, Jain PK (2012) Genetic structure and diversity analysis of the primary gene pool of chickpea using SSR markers. Genet Mol Res 11:891-905
[Crossref] [Google Scholar] [PubMed]
- Dev A, Verma P, Kumhar BL (2017) Genetic character variability studies in desi chickpea (Cicer arietinum L.) genotypes. Int J Curr Microbiol Appl Sci 6:20-25
- Hailu F (2020) Genetic variability, heritability and genetic advance of kabuli chickpea (Cicer arietinum L.) for agronomic traits at Central Ethiopia. J Plant Breed Crop Sci 7:710-714
- Fiaz S, Aslam M, Masood FW, Riaz A, Bashir (2016) Interrelationships among yield and yield contributing traits in chickpea (Cicer arietinum L.). Int J Biosci 9: 49-57
- Holland SM (2016) Principal component analysis. Department of geology, University of Georgia, Athens, GA30602-2501. Indian J Genet Plant Breed 5:415-437
- Joshi P, Yasin M, Sundaram P (2018) Genetic variability, heritability and genetic advance study for seed yield and yield component traits in a chickpea Recombinant Inbred Line (RIL) population. Int J Pure Appl Biosci 6:136-141
- Mayuriben R, Sunayan R, Sunil S, Arpan J, Harshad N (2019) Diversity study through D2 analysis in Chickpea. The Pharma Innovation J 8:140-143
- Millan T, Clarke HJ, Siddique KH, Buhariwalla HK, Gaur PM, et al. (2006) Chickpea molecular breeding: New tools and concepts. Euphytica 147:81-103
- Prakash V (2006) Divergence analysis in Kabuli chickpea (Cicer arietinum L.). Indian J Genet 66:241-242
- Qureshi AS, Shaukat A, Bakhsh A, Arshad M, Ghafoor A (2004) An assessment of variability for economically important traits in chickpea (Cicer arietinum L.). Pak J Bot 36:779-785
- SAS Institute ( 2016) SAS JMP5 Multivariate methods user manual. Inc., Cary, NC, USA.
- Senait Berhanu W (2015) Chickpea (Cicer arietinum L.) fabaceae landrace diversity in Ethiopia. Msc. Thesis, Addis Ababa University, Addis Ababa, Ethiopia.
- Setotaw F, Fikre Asake F, Said A (2018) Assessing the competitiveness of smallholder’s chickpea production in the central highlands of Ethiopia. Ethiop J Crop Sci 6:51–65.
- Sharma JR (1998) Statistical and biometrical techniques in plant breeding. New Age International Publishers, New Delhi 432.
- Singh BD (2001) Plant breeding: Principles and methods. Kalyani Publishers, New Delhi.
- Admas S, Abeje G (2017) Phenotypic diversity studies in chickpea (Cicer arietinum L.) germplasm of Ethiopian collections. Int J Curr Res 9:48506-48512

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences