ISSN : 2476-1974
Reproductive Immunology: Open Access
Friend Leukaemia Integration 1 is Associated with Conception Rate in Holsteins
Mayumi Sugimoto1*, Toshimi Baba2, Yusaku Gotoh2, Takayoshi Kawahara2 and Yoshikazu Sugimoto3
1National Livestock Breeding Center, Nishigo, Fukushima, Japan
2Holstein Cattle Association of Japan, Hokkaido Branch, Sapporo, Hokkaido, Japan
3Shirakawa Institute of Animal Genetics, Nishigo, Fukushima, Japan
- *Corresponding Author:
- Mayumi Sugimoto
National Livestock Breeding Center
Nishigo, Fukushima, Japan
E-mail: m0komats@nlbc.go.jp
Received date: March 04, 2016; Accepted date: April 11, 2016; Published date: April 15, 2016
Citation: Sugimoto M, et al. Friend Leukaemia Integration 1 is Associated with Conception Rate in Holsteins. Reproductiv Immunol Open Acc. 2016, 1:7. doi: 10.4172/2476-1974.100007
Copyright: © 2016 Sugimoto M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Conception rate is an economically important trait in the dairy industry; however, it has decreased dramatically over recent decades. Conception is a complex process including follicle development, ovulation, fertilization, implantation, and placental differentiation and numerous factors contribute to this event. The present study aims to explore the genetics of conception rate in Holsteins using a genome-wide association study (GWAS). Methods and Findings: Our GWAS for conception rate based on 2,559 Holstein sires found that the conception rate is influenced by a single nucleotide polymorphism GA-266del in the 5’ untranslated region of friend leukaemia integration 1 (FLI1). Cows with higher conception rates carried the GA polymorphism in the FLI1 5’ untranslated region. Luciferase assays and quantitative analysis of allele ratios revealed that FLI1 transcripts with the GA polymorphism were expressed at higher levels than those carrying the deletion polymorphism. FLI1 is a member of the ETS gene family of transcription factors and disruption of FLI1 increased natural killer cell population. High levels of natural killer cells were correlated with spontaneous abortion in human. Cows with the deletion polymorphism released higher levels of perforin, a product of natural killer cells, than did cows with the GA polymorphism. Moreover, cows with the deletion polymorphism have lower successful rate for pregnancy after embryo transfer than cows with the GA polymorphism. Conclusions: These observations suggest that cows carrying the deletion polymorphism in FLI1 might have lower conception rates because of the enhanced perforin production. Thus, this study provides novel insights into the role of FLI1 during reproduction process.
Keywords
Conception; GWAS; FLI1; NK cells; Perforin
Introduction
Dairy production depends on the frequency at which cows conceive; thus, conception rate (CR) is essential for this industry. Studying the genetics of CR using a genome-wide association study (GWAS) may be helpful for understanding the underlying biological mechanisms and beneficial for the industry. Here we report a new gene, friend leukaemia integration 1 (FLI1), which is associated with CR in the Holstein cattle population.
FLI1 is a proto-oncogene and belongs to the ETS gene family of transcription factors [1]. The targeted disruption of Fli1 in mice increased numbers of natural killer (NK) cell [2]. High levels of NK cells in the peripheral blood of patients were correlated with spontaneous abortion in human [3]. Activation of NK cells by polynucleotides can cause abortion in pregnant mice [4]. Moreover, the decidual Va14 NKT cells, a subset of NK cells, were involved in abortion through perforindependent pathway [5]. Perforin, a pore-forming protein, acts as a host defence molecule protecting both the mother and the fetus from a wide spectrum of pathogens and also acts as an effector molecule causing the apoptosis of trophoblast cells leading to various pregnancy disorders [6]. Therefore, FLI1 might be correlated with CR through perforin-dependent pathway in ruminants.
In this work, we performed a GWAS on 2,559 Holstein sires and identified a single nucleotide polymorphism (SNP) in the 5’ untranslated region (UTR) of FLI1 that influences the CR of cows. Cows with the deletion polymorphism were less fertile than cows with the GA polymorphism, and this SNP decreased the expression of FLI1. We demonstrated that cows with the deletion polymorphism release more perforin compared with cows with the GA polymorphism. In addition, cows with the deletion polymorphism have lower success rate for pregnancy after embryo transfer than cows with the GA polymorphism. Thus, we propose that FLI1 keeps a low level of perforin that maintains pregnancy.
Methods
Samples
We collected DNA from 2,559 Holstein sires and evaluated the estimated breeding values (EBVs) for the CRs [7,8]. The EBVs for the CRs of the sires were evaluated based on their daughters’ CRs. The EBVs for CRs of daughters were evaluated by threshold linear models using insemination event data after first calving. The threshold-linear repeatability animal model to estimate EBV of CR can be written as:
l=Xβ+Whh+Wss+Zaa+Zpp+e
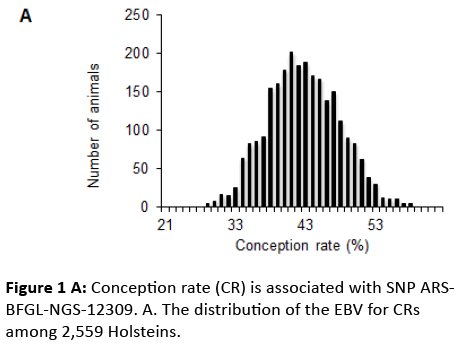
where l is a vector of unobserved liabilities converted from insemination events data, which collected as a longitudinal binary response of either a success or a failure. ; β is the fixed vector of systematic effects (age at insemination, month of insemination, days from calving to insemination, regression coefficients on inbreeding); his the random vector of herdyear- season at insemination; sis the random vector for interaction of service sires and years ; ais the random vector of additive breeding values; pis the random vector of permanent environmental effects; eis the random vector of residual terms; and X, Wh, Ws, Za, and Zp are known incidence matrices with the appropriate dimensions. The EBV for the CRs of the population was distributed as shown in Figure 1A.
Whole-genome scan
We genotyped 2,559 samples using a Bovine SNP 50 v1 DNA Analysis Kit ( Illumina, San Diego, CA, USA) for a total of 54,035 SNPs and conducted an association analysis using EMMAX software [9].
Identification of novel SNPs
Based on the Nov. 2009 Bos taurus draft assembly [10] (UMD_3.1), each of the exons, 2 kb of the 5’UTRs and 2 kb of the 3’UTRs of the genes located in the associated regions were amplified by polymerase chain reaction (PCR) and sequenced. The genome-wide regions that included significant SNPs as well as their neighbouring SNPs with r2 values greater than 0.2 were defined as the associated regions. The r2 values were calculated by a linkage disequilibrium analysis using PLINK software [11]. The primers for each gene and the samples used to compare the sequences are shown in Tables 1 and 2, respectively.
| Gene | Position | Primer | Sequence |
|---|---|---|---|
| ETS1 | 5'UTR-1 | Forward | GTGGTTAGCAGTGTTTAGGCT |
| Reverse | ACACACCTGCTTACCTCATCT | ||
| 5'UTR-2 | Forward | TTCTCTTCCTGGCTCCTTCC | |
| Reverse | TGTCACCACTGGCCAAAATT | ||
| 5'UTR-3 | Forward | CAGAGCTGTGCATCATGTTTT | |
| Reverse | TCCACGCATTCTTGAGGACT | ||
| 5'UTR-4 | Forward | AGCAGCCCAAGTCCAGTATT | |
| Reverse | GGGGAATCGGACCTTCTTCT | ||
| exon 1 | Forward | AGAGATCCTGAGGGTGGGG | |
| Reverse | GGGGAGAAGTGGAGGGGA | ||
| exon 2 | Forward | GCAGAACGATCACCACCATC | |
| Reverse | GGTCCATCCTCTCTCCTTCC | ||
| exon 3 | Forward | TTTCGTGTAGTCTCCGAGGC | |
| Reverse | CACCCTGTCCTCATGCATTT | ||
| exon 4 | Forward | TGAGATCACTGTGGTCCTCG | |
| Reverse | GGAAGAGAGAAGAGGAGCCA | ||
| exon 5 | Forward | TCTCCTCTCTCCAATCGCAC | |
| Reverse | TGGCTAAGAGTGAGGGAGGA | ||
| exon 6 | Forward | GTGTCTCTTCCCATCCCTCC | |
| Reverse | CAGAAGTGTCCAGGGAGCC | ||
| exon 7 | Forward | TTCACCATGGCTTGTGTCTC | |
| Reverse | ACACCATCAAGCCCCATACA | ||
| exon 8 | Forward | CACCAATGAGTGCAGGCATA | |
| Reverse | TCAGAATCCTCAGTCGGCAA | ||
| 3'UTR-1 | Forward | GATGGACTTCAGTGGGGAGG | |
| Reverse | ATAAATGTGGGGTGCTGGGA | ||
| 3'UTR-2 | Forward | GGAAAGAGGGAGTGAAGGGA | |
| Reverse | TTGACAATGGCCTCGGTTTG | ||
| 3'UTR-3 | Forward | AAGGAAGGAAGCTTGAAGGC | |
| Reverse | CTCCCTGAGCAGCTCCTAAA | ||
| 3'UTR-4 | Forward | GCCCAGCTGTGTATTGTGAT | |
| Reverse | TCTTCCTGGGATGGTCTCTG | ||
| FLI1 | 5'UTR-1 | Forward | GAGGAAAGGGTTAAGCCTGATT |
| Reverse | CTTCTTTCTCCCCGACTTCC | ||
| 5'UTR-2 | Forward | AAAGTCCAAAGCGTGGTCTG | |
| Reverse | TGCATCCAATGGGAAGTTTT | ||
| 5'UTR-3 | Forward | GAGCTCTCCAGTAGCCCAGA | |
| Reverse | TTGTTCCCGGGAGATAAGG | ||
| 5'UTR-4 | Forward | TGCAGACTTTGGGAATCAGG | |
| Reverse | GCGGAAGGAAGGGAGAGT | ||
| exon 1 | Forward | CTTTTTCGCTCCGCTACAAC | |
| Reverse | GCGGAAGGAAGGGAGAGT | ||
| exon 2 | Forward | GGGCTCTGTGTCCTTCTCTG | |
| Reverse | CGTCTGCCACAGACACACTT | ||
| exon 3 | Forward | CCTTCCCCTGAGCTTTGTCT | |
| Reverse | AGTGAAAGGGTTCCCGAAGT | ||
| exon 4 | Forward | TTGCTAACAGCCTGTCTCTCC | |
| Reverse | TAGGGACCGGGCACTTAC | ||
| exon 5 | Forward | GTTTTTGCTTCGCCTTTCAG | |
| Reverse | CCCAGTCTTCCCATCACAGT | ||
| exon 6 | Forward | CTGCCACTCCATGAGCTGTA | |
| Reverse | CGCTTATGACCCTGTTCTCC | ||
| exon 7 | Forward | GGGAGTGAGTGAATGGGAAA | |
| Reverse | AGGGTTCGAACATCATGGAC | ||
| exon 8 | Forward | TCAGGCTTTCCTATGATCTCAA | |
| Reverse | ACACAACCTCTCAGGCCAAA | ||
| exon 9 | Forward | TCTCAGGTGGAGCCTGTTTT | |
| Reverse | CCACCGATGAGGAAGCAT | ||
| 3'UTR-1 | Forward | CATCTACCCCAACCCCAAC | |
| Reverse | GTTCCAGTTGCCCTCCACT | ||
| 3'UTR-2 | Forward | GGCAGGAAGCTTATCATCTTATC | |
| Reverse | AACGTACAAGCAGCCCAAAT | ||
| 3'UTR-3 | Forward | GAGTTGACCTCGGTCACAGAT | |
| Reverse | CTGGGAAAACCCTTGGACCT | ||
| 3'UTR-4 | Forward | TTGTGCCTTCTTCTCTCAGAAC | |
| Reverse | CTACACCATCAGCCGGTTTC | ||
| KCNJ1 | 5'UTR-1 | Forward | AGGCTGGTCTGAGGGACAAT |
| Reverse | CCTTTCTCCCTGGCTTTACC | ||
| 5'UTR-2 | Forward | AAAGAAAAGCCTTCCATGAGC | |
| Reverse | CTCCAGTCAGTGCAGAACCA | ||
| 5'UTR-3 | Forward | TGCAAATGAATGAGGCACTT | |
| Reverse | TGGCATTGAGTGACTGTTCC | ||
| 5'UTR-4 | Forward | TGTGTGAGCCAGAGATGACC | |
| Reverse | TCAGACCAGCTGCCAACTC | ||
| exon 1 | Forward | TGTATCCTGCCCACTTACCC | |
| Reverse | TATGGCATTTCTCCGCTTAC | ||
| exon 2 | Forward | CTTCTCTTAGTGACTTTCTGTTCTGA | |
| Reverse | CCCCTGTCCTGTGATGAATG | ||
| exon 3 | Forward | TCTGTTTTGTCTTTCTCTGATGTGT | |
| Reverse | ACTCGTGTGGAAAACTCAGC | ||
| 3'UTR-1 | Forward | TGAAACAGACGACACCAAAA | |
| Reverse | GAGCCAATGTTCAAATAAAAGTGA | ||
| 3'UTR-2 | Forward | ATGGACAGGCCAAATGAGAT | |
| Reverse | TGCCTTCTTGGAAGATCAGC | ||
| 3'UTR-3 | Forward | CGCTGGCTTCAAATCTGTTA | |
| Reverse | GGACGTCACAACGTCAGAGA | ||
| 3'UTR-4 | Forward | GTATTCTGGAGCGGACGGTA | |
| Reverse | TCATAGCAATGGGTCAGCAG | ||
| ARHGAP32 | 5'UTR-1 | Forward | CTGTGCCTCTCATTGTGCTG |
| Reverse | TGAGTTGTATGAGCTACTTGTGT | ||
| 5'UTR-2 | Forward | ACAGAATGGGAGGAAATATTTGC | |
| Reverse | TCATCTCTAGCTCCATCCATGT | ||
| 5'UTR-3 | Forward | ATCCTGTAGTGCCACTCCTG | |
| Reverse | GACTGTGAACCAATCCAACATAA | ||
| 5'UTR-4 | Forward | ACTTCAACTTAAGGGGAACGTG | |
| Reverse | CGAATCCAACCAGAACACG | ||
| exon 1 | Forward | GAACAACCTGGACACTGCTG | |
| Reverse | AGATGAGGGAGGTGGAGAGA | ||
| exon 2 | Forward | TGGATTGTAGTCATTGGAAGGC | |
| Reverse | TGCTTCCCCTGTTCCTTTTC | ||
| exon 3 | Forward | TCTTTTGGTATGGAGTTAGGACC | |
| Reverse | TGTATGGACAGTAAGAGCTCATT | ||
| exon 4 | Forward | TGTTCCAGGATCTTGTCTCTCA | |
| Reverse | ACGCCTCGCTTCAGTATGTA | ||
| exon 5 | Forward | ACCGTGACTTTCTTTCCCTCT | |
| Reverse | TGCAGAAATGCCAATGTGACT | ||
| exon 6 | Forward | TCGCTAGAGGGTTTTGGAGT | |
| Reverse | CCCATGAATCTTCCTGAGTGC | ||
| exon 7 | Forward | TCTGTGAAGAACCTCTGTGACT | |
| Reverse | CACATGGGATTTCTTTCGTAGGT | ||
| exon 8 | Forward | CTTGCAAGTCCCGTGTCTTT | |
| Reverse | GCTAGAAAGGCAGCACAGAC | ||
| exon 9 | Forward | GACCAGTCTTTGTGCTGCTT | |
| Reverse | CACACCGAATTCTTTGTTGCG | ||
| exon 10 | Forward | GCTCACACACTGGTCTGTTC | |
| Reverse | TGACAACGAACACAACAGCT | ||
| exon 11 | Forward | TGCACTTCATTTCCTCTTGCT | |
| Reverse | GGAGATCAACAGGGAGAGGT | ||
| exon 12 | Forward | TTAATCTCCACCCCTTACAGC | |
| Reverse | GAGGCCAAGGTTTTCTGATACT | ||
| exon 13-14 | Forward | CTCACTGAGCTGGAGGTTAAT | |
| Reverse | TGGCATTTTACAGAGCGTGG | ||
| exon 15-16 | Forward | ACTCCCTGATGTTTCTTTGTGT | |
| Reverse | TTAAAGATACGTTCCCAGGGG | ||
| exon 17 | Forward | AGTTTGAGTCTTGTCGTTGCT | |
| Reverse | CCACAGTCGAATGAACAGGT | ||
| exon 18 | Forward | TTGGATGCCTTAATGCGGAC | |
| Reverse | AAGGGAAGGCGTGAACACT | ||
| exon 19 | Forward | ACGTTGTTTGGTTTTGATTCTGC | |
| Reverse | CGTAGTGACTGAACAACAACAAC | ||
| exon 20 | Forward | AGCTGACTCATAGGGCACTG | |
| Reverse | AAGCAAGAATGGAGGAAAACAAA | ||
| exon 21-1 | Forward | ACACAAGCTACCTTTTCACTTTC | |
| Reverse | GGCACACTGGTCTTCACAGA | ||
| exon 21-2 | Forward | CAGAGTCACTTCCGTTCCCT | |
| Reverse | GGCACACACTGATGGAGAGT | ||
| exon 22-1 | Forward | ATTCCGCCTGCAGTCCAT | |
| Reverse | GGTCTGGAAGCCCTTTGG | ||
| exon 22-2 | Forward | GGGCAGAGTATGTGTCCTCA | |
| Reverse | GGAGTCCAGTTTTCCCAGGA | ||
| exon 22-3 | Forward | GAGAAATACCGCCTGCAGTC | |
| Reverse | CCTCCAGGTTATCGTACTGC | ||
| exon 22-4 | Forward | GGGTCACCTGTTTTCTTTGTCA | |
| Reverse | ACTGAAGTGTGTGGAGCAAC | ||
| 3'UTR-1 | Forward | GACCCATTAGATCCAGGCTGA | |
| Reverse | GCTGGTGTGGATGGCAATTA | ||
| 3'UTR-2 | Forward | GGAATTACCGTGTTGTGTCTTC | |
| Reverse | ACACCTAACAGAGTATTTCCACA | ||
| 3'UTR-3 | Forward | TCATGTGATTGCATTTTAAGGGT | |
| Reverse | ACGTTTCACACTTTCACCAGG | ||
| 3'UTR-4 | Forward | GGTCACACACACTGTTTACTCC | |
| Reverse | TACTGAAAGGAGGCGGCATC | ||
| JAM3 | 5'UTR-1 | Forward | GCATAGACTCCACAGCCCTA |
| Reverse | CAGCCTCTGTCCCATAAACA | ||
| 5'UTR-2 | Forward | AAGTCAGCGGGCCTAAGTAG | |
| Reverse | TGCTCCAGGAAACAACAAACT | ||
| 5'UTR-3 | Forward | TGACTGTGTGAAAAGTGACGT | |
| Reverse | GAACCCGGGTCTCCTTCAT | ||
| 5'UTR-4 | Forward | TTCAGCACTTTCCCCTCTCA | |
| Reverse | CCTCAGCGCCATGTCGAG | ||
| exon 1 | Forward | TCCATAGCAACCAGACTCGG | |
| Reverse | CGAGACCCTTCCCTGACG | ||
| exon 2-3 | Forward | CGTCTCTGACTTGGCTTTTCT | |
| Reverse | GGGCCTTCTGTACAAAGAGG | ||
| exon 4-5 | Forward | TCCTTTAACGGGGAAGCCTT | |
| Reverse | GGTCTGTAGCTCTGGTCTCC | ||
| exon 6 | Forward | ACATTGTTGGTGTGTTCGGG | |
| Reverse | GAGGCTCAGCAGACACTCA | ||
| exon 7-8 | Forward | ACAGCATCTTCTCACCCCTC | |
| Reverse | CGTCTCCAGGCTCCCTTAC | ||
| exon 9-1 | Forward | ATCGAGGGAAGTGGTGTCAG | |
| Reverse | GGTTTCCTAAGCCACCAGTG | ||
| exon 9-2 | Forward | TGTTCTGCTTTTCTATGGGTGT | |
| Reverse | TGTCTTCATGGCAGAGGGAC | ||
| exon 9-3 | Forward | AGGAAAAGGCTACCCACTCC | |
| Reverse | GAAAGAAACTGGGCTGGCTC | ||
| exon 9-4 | Forward | AAAAGGCTACCCACTCCAGT | |
| Reverse | AAAGAAAGGTCAACACAGTCTTG | ||
| exon 9-5 | Forward | ATGGTCCAGGGCCAAAGG | |
| Reverse | AATGAAGAGGCTGAGCTGCT | ||
| exon 9-6 | Forward | TGCCATGAGAACTGGTAGCA | |
| Reverse | GACCCACACTCACTCCTCTC | ||
| 3'UTR-1 | Forward | GCTACTAACACACCTGCACG | |
| Reverse | GGGACCCAAGCTTTGTTTCA | ||
| 3'UTR-2 | Forward | CTCCCGTTGCTCTGGTAAAA | |
| Reverse | TGGTGCTCAGAAAGTGGTCA | ||
| 3'UTR-3 | Forward | AGGGAGAGAAGCTGGGAGTA | |
| Reverse | GCGGTTTCCAAGGTACATCC | ||
| 3'UTR-4 | Forward | GGGATGATTGTACATGTGCAGT | |
| Reverse | CTCTGCTACCCGACTGAAGG | ||
| SPATA19 | 5'UTR-1 | Forward | TGTGCAGATTTAGCGCTTATG |
| Reverse | CTGTTTCCATCCTTACTGGTG | ||
| 5'UTR-2 | Forward | CAAGGTTGAAGAGACAGGGAAG | |
| Reverse | CTCCCCAGCTTCCCTAAAAT | ||
| 5'UTR-3 | Forward | GCCTAGCATATTCTGATCAATAGAGA | |
| Reverse | CACAAGTGATTCAGATCACAAAAA | ||
| 5'UTR-4 | Forward | TCCTTCACAAGAATTGGCACT | |
| Reverse | TCCATTGAAGCAGCCTGAG | ||
| exon 1 | Forward | CCAATCAGGTAGGCACCAC | |
| Reverse | CTCTCCACCTAGCCATCACC | ||
| exon 2 | Forward | TGGATGTGGATAGTGGAGCA | |
| Reverse | TGATGAATCAGACGCAGCTC | ||
| exon 3 | Forward | GGACCAGACGAGTGAAGGAA | |
| Reverse | CGGCTAACAGGCTCCATTAC | ||
| exon 4 | Forward | CTTGCTGGGCAGTAACCACT | |
| Reverse | GGTTTGTGTCAGCCAGGAAT | ||
| exon 5 | Forward | ATCCCAATGCTTGACGATGT | |
| Reverse | GACCCACAGAGACCAGAAGC | ||
| exon 6 | Forward | CTGCTGTGGTCTGTGCTCTG | |
| Reverse | CCCCTTGCTGGTATTCTCAA | ||
| exon 7 | Forward | GCATTTCCCTCTTTCCATGT | |
| Reverse | TAGCAAGGCACTCACACCAG | ||
| 3'UTR-1 | Forward | AGTGGCAGCACTGGAATCTT | |
| Reverse | TGTGTAAGGAAAGGACGCACT | ||
| 3'UTR-2 | Forward | GTGACATGTGCGCTTCACTC | |
| Reverse | AAGATCTGGGGGACAATTCC | ||
| 3'UTR-3 | Forward | TTGTTCCTCTCCTGGTTTCC | |
| Reverse | ACAGCTGCCGGAAAGAGTAA | ||
| 3'UTR-4 | Forward | GGCCTTCATCGTGAGGTTTA | |
| Reverse | CTGCAGGAATCCTTCCAGAC |
Table 1: Primers used to search for SNPs.
| ID | 34 | 55 | 109 | 152 |
| Paternal ID | 2182318 | 2183007 | 2265005 | 2290977 |
| Maternal ID | 769202561 | 15937840 | 123597843 | 128824973 |
| EBV | 0.521 | 0.4807 | 0.2988 | 0.2937 |
| Hapmap51018-BTA-65434 | G | G | G | G |
| G | G | G | G | |
| Hapmap34976-BES2_Contig422_801 | G | A | G | G |
| A | A | A | G | |
| BFGL-NGS-112928 | G | G | G | G |
| G | G | A | G | |
| ARS-BFGL-NGS-70760 | C | C | A | A |
| C | C | A | A | |
| BTA-65424-no-rs | A | A | A | A |
| A | A | A | A | |
| BFGL-NGS-109714 | A | A | A | A |
| G | G | G | A | |
| ARS-BFGL-NGS-62183 | C | A | A | A |
| C | A | A | A | |
| BTB-01020010 | G | A | A | G |
| G | G | G | G | |
| BFGL-NGS-110219 | A | A | G | G |
| G | G | G | G | |
| Hapmap40017-BTA-65421 | C | C | A | A |
| C | C | C | A | |
| ARS-BFGL-NGS-36127 | A | A | A | A |
| A | A | A | A | |
| UA-IFASA-7281 | G | G | A | A |
| G | G | A | A | |
| ARS-BFGL-NGS-87575 | G | G | A | A |
| A | G | A | A | |
| ARS-BFGL-NGS-12309 | G | G | A | A |
| G | G | A | A | |
| BTA-65417-no-rs | G | G | A | G |
| G | G | G | G | |
| UA-IFASA-7219 | A | G | A | A |
| A | A | A | A | |
| ARS-BFGL-NGS-1333 | A | A | G | G |
| A | A | G | G | |
| BTA-65410-no-rs | A | G | G | G |
| G | G | G | G | |
| UA-IFASA-8767 | G | A | A | A |
| G | A | A | A | |
| UA-IFASA-9430 | A | G | A | A |
| A | A | A | A | |
| Hapmap48423-BTA-65404 | A | G | A | A |
| A | A | A | A | |
| BFGL-NGS-119359 | G | G | G | G |
| G | G | G | G | |
| ARS-BFGL-NGS-16031 | A | A | G | G |
| A | A | G | A | |
| Hapmap58618-rs29012371 | G | G | A | G |
| G | A | A | A | |
| BTA-65537-no-rs | A | A | A | A |
| G | G | G | A | |
| BFGL-NGS-117839 | G | G | G | |
| A | A | A | ||
| BTB-01023253 | A | G | A | A |
| G | G | A | G | |
| ARS-BFGL-NGS-102550 | A | G | A | A |
| A | A | A | A | |
| ARS-BFGL-NGS-24769 | G | G | G | G |
| G | G | G | G | |
| ARS-BFGL-NGS-41631 | A | A | A | A |
| A | A | A | A | |
| BFGL-NGS-115193 | A | A | A | A |
| G | G | G | A | |
| Hapmap52938-rs29027128 | A | A | A | A |
| A | A | A | A | |
| ARS-BFGL-NGS-40178 | G | G | G | G |
| G | A | G | G | |
| BTA-65587-no-rs | A | A | A | A |
| A | A | A | A | |
| UA-IFASA-8226 | G | A | A | A |
| A | A | A | A |
Table 2: Samples used for developing new SNPs.
Allelic substitution effects
We genotyped 2,682 cows and 4,165 bulls for FLI1, PKP2, CTTNBP2NL, SETD6, CACNB2, UNC5C, and FAM213A. FLI1 was identified as a gene that was associated with the CR in the present study. PKP2, CTTNBP2NL, SETD6, CACNB2 and UNC5C have previously been identified as genes associated with the CR, whereas FAM213A has previously been identified as a gene that was associated with the fertility selection index (SI) [12-14]. The SI consists of the EBVs for days open (DO), the number of inseminations per lactation (NI), success after first insemination (SFI), and pregnancy within 70 d (P70), 90 d (P90), and 110 d (P110) after delivery [15]. The EBVs of cows and bulls with these six traits included in the SI were estimated by an animal model using 1,881,898 records. The EBVs of cows and bulls in relation to the CR were estimated by a thresholdlinear repeatability animal model using 3,428,666 values after first parturition. The data were collected between January 1990 and September 2015 by the dairy herd improvement program of Hokkaido, Japan. The allelic substitution effects of these genes were determined using the following equation:
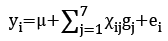
where yi = the deregressed EBV [16] of animal i (= 1, 2, …, n) for the CR, DO, NI, SFI, P70, P90, P110 or SI; μ = the general average value of the population; χij= the genotype covariate (coded as 0 or 2 for the two homozygotes and 1 for heterozygotes) of gene j in animal i; gj = the random regression coefficient representing the allelic substitution effect for gene j; and ei = the random residual effect for the value of animal i. We performed the analyses with the SAS MIXED procedure.
Real-time PCR
RNA was extracted from individual samples of bovine brain, heart, kidney, liver, lung, ovary, pancreas, skeletal muscle, spleen, stomach, and uterine tissue using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Real-time PCR was conducted with an ABI 7900HT Sequence Detection System using the comparative Ct method and glyceraldehyde-3- phosphate dehydrogenase (GAPD) as an internal control (Life Technologies). The primers used in these assays are shown in Table 3.
| Gene | Primer | Sequence |
|---|---|---|
| FLI1 | Forward Reverse Probe | CGTCAAGCGCGAGTACGA TGCCGCATTTGCTGACACT TGGGTCCAGGGAGTCTCCGGTG |
| GAPD | Forward Reverse Probe | GCCCTCAACGACCACTTTGT CCTGTTGCTGTAGCCAAATTCA AAGCTCATTTCCTGGTACGA |
Table 3: Primers used for real-time PCR.
Luciferase assay
Fragments of the 5’UTR of FLI1 were generated using PCR with the respective forward and reverse primers (Table 4). These PCR products were further amplified via PCR using the forward2 and reverse2 primers (Table 4) to be cloned into a pGL3 (R2.2)-basic vector (Promega, Madison, MI, USA) using an In-Fusion Advantage PCR Cloning Kit (Takara Bio Inc., Shiga, Japan). Luciferase assays were performed using a Dual- Luciferase Reporter Assay System (Promega).
| Primer | Sequence |
|---|---|
| Forward | TATATAGTGTGTGTGATGCG |
| Reverse | TTGGCCAAGTCTGCAGCCGA |
| Forward2 | CCGAGCTCTTACGCGTTATATAGTGTGTGTGA |
| Reverse2 | CTTAGATCGCAGATCTTTGGCCAATCTGCAG |
Table 4: Primers used for generating reporter constructs.
SNaPshot and quantitative analysis of allele ratios
The allelic messenger RNA (mRNA) ratio was determined using a SNaPshot Multiplex Kit (Life Technologies), and the primers used are shown in Table 5. For cDNA preparations, each mRNA was converted to cDNA in three separate experiments.
| Primer | Sequence |
|---|---|
| Forward | TATATAGTGTGTGTGATGCG |
| Reverse | CTTTGCGAATGGGGAGGAAG |
| Extension | GCGCGAGACAGAGAGAGAGAGAGAGAGAGAGA |
Table 5: Primers used for SNaPshot.
Enzyme immunoassays
The concentration of perforin released from the bovine serum was assayed using a Perforin Human ELISA Kit (Abcam, Cambridge, UK) according to the manufacturer’s instructions.
Embryo transfer to recipient female
The embryos were transferred non-surgically into Holstein heifers to the uterine horn ipsilateral to the existing corpus luteum using an embryo transfer device (Misawa Medical Industry Co., Ltd, Tokyo, Japan) on days 6–8 of the estrous cycle. Pregnancy was determined by real-time B-mode ultrasonography (Honda electronics Co. Ltd., Toyohashi, Japan) on days 30 and 60 of gestation. Calves were delivered spontaneously without induction of parturition.
Results
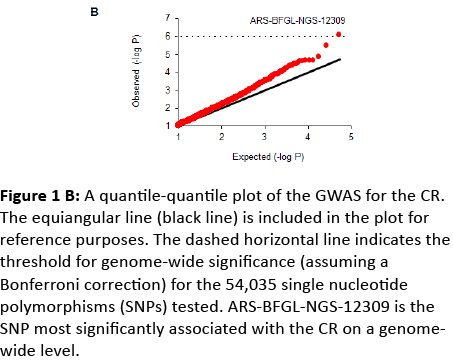
Our GWAS identified ARS-BFGL-NGS-12309 as the most significantly associated SNP on chromosome 29 (Figure 1B), and the region associated with this SNP included 6 genes (Figure 2A). To detect possible causative polymorphisms in this region, we sequenced all exons and the 5’ and 3’ UTRs of these 6 genes and found 35 novel SNPs (Figure 2A). A reanalysis of the newly sequenced SNPs demonstrated that FLI1 (GA-266del) was the most significant (Figure 2A). Moreover, we genotyped FLI1 (GA-266del) in 2,682 cows and 4,165 bulls and found that the allele substitution effect of FLI1 on the dEBV for the CR was 0.44 (Figure 2B). Cows with the GA/GA genotype exhibited a 0.44 higher CR than those with the GA/del genotype. FLI1 also had favourable effects on the dEBV for the traits that compose the SI (DO, NI, SFI, P90, and P110) and the SI itself. The effects of FLI1 were similar to those of PKP2, SETD6, CACNB2, UNC5C, or FAM213A, which have previously been identified to be associated with the CR or SI in the Japanese dairy cow population [12-14] (Figure 2B). Therefore, FLI1 (GA-266del) was the most promising causative SNP on chromosome 29 and had a similar impact on the CR as the five genes previously identified to be associated with CR or SI.
Figure 1b: A quantile-quantile plot of the GWAS for the CR. The equiangular line (black line) is included in the plot for reference purposes. The dashed horizontal line indicates the threshold for genome-wide significance (assuming a Bonferroni correction) for the 54,035 single nucleotide polymorphisms (SNPs) tested. ARS-BFGL-NGS-12309 is the SNP most significantly associated with the CR on a genomewide level.
Figure 2: FLI1 SNP is associated with the CR. A. Association signals on chromosome 29 with the CR using plots of the P values from the EMMAX analysis. The black line represents the threshold for genome-wide significance after applying the Bonferroni correction for multiple comparisons. ARSBFGL- NGS-12309 was the first significantly associated SNP that was detected, whereas FLI1 (GA-266del) was the most significantly associated novel SNP. Blue circles represent the positions of the 35 newly sequenced SNPs. Blue lines represent 6 genes located in the associated region. The triangle diagram represents pairwise linkage disequilibrium (LD) in the associated region, which was visualized using Haploview [17]. Red shades indicate strong LD. B. The allelic substitution effects of FLI1, PKP2, CTTNBP2NL, SETD6, CACNB2, UNC5C, and FAM213A on the deregressed EBV [16] for CR, days open (DO), number of inseminations per lactation (NI), and success after first insemination (SFI), pregnancy within 70 d (P70), 90 d (P90), and 110 d (P110) after delivery or fertility selection index (SI). n. s.: nonsignificant. * and **: p < 0.05 and p < 0.01, respectively.
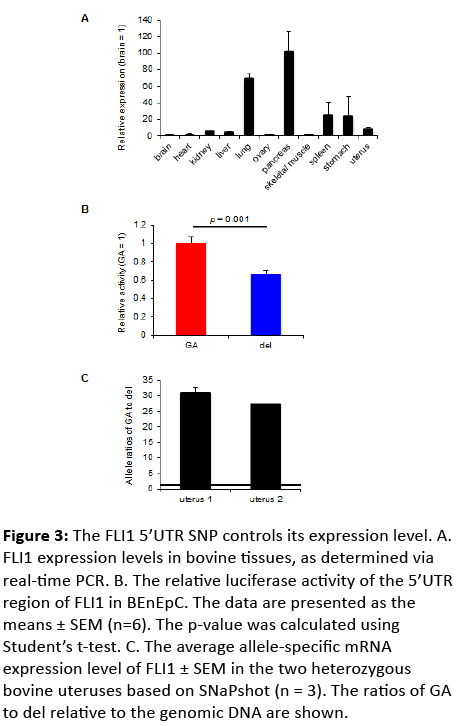
FLI1 (GA-266del) is located in the 5’UTR of FLI1 and may influence the expression level of this gene. Because FLI1 is expressed in several bovine tissues, including the uterus (Figure 3A), we used BEnEpC derived from bovine uterine tissue to assess luciferase activity. Reporters carrying the GA allele exhibited higher activity than those carrying the del allele (Figure 3B). Consistent with the results of the luciferase assay, the level of mRNA generated in the presence of the GA allele was higher than that produced in the presence of the del allele according to the allelic mRNA ratio measured in the bovine uteruses (Figure 3C). Consequently, the FLI1 expression level might affect CR in cattle.
Figure 3: The FLI1 5’UTR SNP controls its expression level. A. FLI1 expression levels in bovine tissues, as determined via real-time PCR. B. The relative luciferase activity of the 5’UTR region of FLI1 in BEnEpC. The data are presented as the means ± SEM (n=6). The p-value was calculated using Student’s t-test. C. The average allele-specific mRNA expression level of FLI1 ± SEM in the two heterozygous bovine uteruses based on SNaPshot (n = 3). The ratios of GA to del relative to the genomic DNA are shown.
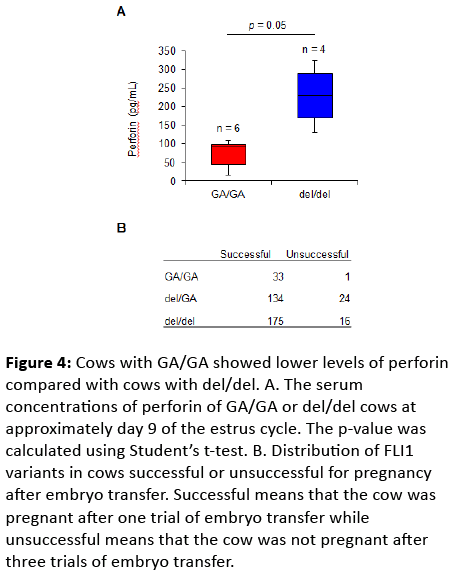
The reduced expression of Fli1 in mice increased numbers of NK cell [2] and NK cells were involved in the acceptance of the fetus to the mother through perforin-dependent pathway [5]. Thus cows with the reduced expression of FLI1 might show high concentration of perforin and low CR. To explore the possibility, we examined the FLI1 genotypes and their perforin concentration in cows. As expected, the serum concentrations of perforin of cows carrying del/del were higher than those of cows carrying GA/GA (Figure 4A). Moreover, we found a relationship between the FLI1 genotype and outcome of embryo transfer (Figure 4B). 33 cows carrying GA/GA were pregnant after one trial of embryo transfer (Successful) while only 1 cow carrying GA/GA was not pregnant after three trials of embryo transfer (Unsuccessful). On the other hand, 16 out of 191 cows carrying del/del were unsuccessful. The chi-square statistic for the distribution of FLI1 variants gives a p-value of 0.038. Therefore, the reduced expression of FLI1 increased perforin production, which might decrease CR through inhibiting to accept the embryo to the recipient.
Figure 4: Cows with GA/GA showed lower levels of perforin compared with cows with del/del. A. The serum concentrations of perforin of GA/GA or del/del cows at approximately day 9 of the estrus cycle. The p-value was calculated using Student’s t-test. B. Distribution of FLI1 variants in cows successful or unsuccessful for pregnancy after embryo transfer. Successful means that the cow was pregnant after one trial of embryo transfer while unsuccessful means that the cow was not pregnant after three trials of embryo transfer.
Discussion
The present study is the first to demonstrate that FLI1 modulates CR. Analyzing the whole genome of 2,559 Holstein sires identified a novel mutation associated with the CR. Although we have previously identified several genes associated with the CR in the Japanese Holstein female population [12,13], FLI1 is a novel gene associated with CR, which has previously been known as a proto-oncogene and belongs to the ETS gene family of transcription factors [1]. One of the reasons for this GWAS result might be that we scanned the whole genome of sires whose EBVs for traits are more precise than females because of their large number of offspring. A second reason might be that we analyzed all of the 2,559 samples rather than comparing two groups of samples carrying the extremes of CR.
We found that the SNP in the 5’UTR of FLI1 is correlated with CR. The region including the SNP identified is not predicted to be a binding site of transcription factor (TRANSFAC 7.0, https://www.gene-regulation.com/pub/datab ases.html), however, it might affect interaction with an unknown nuclear protein [18]. cis-Acting polymorphisms would affect transcription, mRNA processing, mRNA stability, and protein translation [19]. Several GWAS reported that SNPs located in 5’UTR of the genes were associated with a broad range of phenotypes [20-22]. Since the polymorphism in the 5’UTR of FLI1 influences its expression level (Figures 3B and 3C), the associated genetic variant should harbor the functional effect and affect the phenotype.
Several studies implicate that oncogenes play an important role in fertility [23,24]. One example is pleomorphic adenoma gene 1 (PLAG1) known as an oncogene associated with pleomorphic adenomas of the salivary gland, which belongs to the PLAG family of zinc finger transcription factors [25]. The study of Plag1 knockout mice suggests that PLAG1 deficiency causes growth retardation as well as reduced fertility [26]. GWAS in humans and domestic animals indicated that polymorphisms in the PLAG1 genomic region were associated with body growth and reproductive traits [27,28]. Possible mechanisms linking PLAG1 to reproductive physiology could be related to growth hormone (GH) and insulin-like growth factor (IGF) 1 or 2 signalling [24]. Interestingly, IGF1 is a target gene of FLI1 [29]. Moreover, administering of bovine somatotropin to dairy cows increased plasma concentrations of GH and IGF1 and enhanced conceptus size and fertility [30]. FLI1 might influence CR through GH and IGF1/2 signalling as well as PLAG1.
We demonstrated here that the polymorphism in FLI1 affected the serum concentration of perforin (Figure 4A). In cattle, perforin was highly expressed in the peri-implantation endometrium, suggesting that it may play important roles in the establishment and maintenance of gestation during normal pregnancy in ruminants [31]. By producing perforin, uterine NK cells act as a double-edged sword at the maternalfetal interface to protect the host from the pathogens and along with apoptosis of fetus [6]. In dairy cattle, infection of the mammary gland is associated with a reduction in pregnancy rate and an increase in the number of inseminations required to establish pregnancy [32], suggesting that activation of the immune system by infection inhibits pregnancy. Keeping the appropriate level of perforin would be critical for normal pregnancy in ruminants as well as other mammals.
Several reports inferred the link between immunity and female fertility. The long pentraxin 3 is produced by innateimmunity cells in response to proinflammatory signals and acts as a predecessor of antibodies, but it is also essential in female fertility by acting as a nodal point for the assembly of the cumulus expansion [33]. Peroxisome proliferator-activated receptor gamma regulates immune cell activation and facilitates the release of oocytes each estrous cycle [34]. FLI1 might have dual roles for controlling NK cell population and CR.
In conclusion, the present study investigated the whole genome in 2,559 samples, and a significant association was observed between CR and FLI1 with a polymorphism in 5’UTR. Further functional studies revealed that this SNP was correlated with the expression of FLI1, the concentration of perforin, and the outcome of embryo transfer. These results indicate that FLI1 plays a role in female fertility in cows.
Acknowledgments
The authors thank K. Maruyama for performing extensive genotyping.
Funding
This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Research Program on Innovative Technologies for Animal Breeding, Reproduction, and Vaccine Development, AGB-2004).
References
- Watson DK, Smyth FE, Thompson DM, Cheng JQ, Testa JR, et al. (1992) The ERGB/Fli-1 Gene: Isolation and characterization of a new member of the family of human Etstanscription factors. Cell Growth Differ 3: 705-713.
- Masuya M, Moussa O, Abe T, Deguchi T, Higuchi T, et al. (2005) Dysregulation of granulocyte, erythrocyte, and NK cell lineages in Fli-1gene–targeted mice. Blood 105: 95-102.
- Perricone C, De Carolis C, Giacomelli R, Zaccari G, Cipriani P, et al. (2007) High levels of NK cells in the peripheral blood of patients affected with anti-phospholipid syndrome and recurrent spontaneous abortion: a potential new hypothesis. Rheumatology (Oxford) 46: 1574-1578.
- Kinsky R, Delage G, Rosin N, Thang MN, Hoffmann M, et al. (1990) A murine model of NK cell mediated resorption. Am J ReprodImmunol 23: 73-77.
- Ito K, Karasawa M, Kawano T, Akasaka T, Koseki H, et al. (2000) Involvement of decidual Va14 NKT cells in abortion. ProcNatlAcadSci 97: 740-744.
- VeljkovicVujaklija D, Sucic S, Gulic T, Dominovic M (2012) Cell death mechanisms at the maternal-fetal interface: insights into the role of granulysin. ClinDevImmunol 12: 180-272.
- Kawahara T, Gotoh Y, Masuda Y, Yamaguchi S, Suzuki M (2010) Influences of genetic and environmental factors on cow conception rate of Holsteins in Japan using longitudinal binary data. Nihon ChikusanGakkaiho 81: 121-132.
- Averill TA, Rekaya R, Weigel K (2004) Genetic analysis of male and female fertility using longitudinal binary data. J Dairy Sci 87: 3947–3952.
- Kang HM, SulJH, Service SK, Zaitlen NA, Kong SY, et al. (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42: 348-354
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, et al. (2009) A whole-genome assembly of the domestic cow, Bostaurus Genome Biol. 10: 42.
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 81: 559-575.
- Sugimoto M, Sasaki S, Gotoh Y, Nakamura Y, Aoyagi Y, et al. (2013) Genetic variants related to gap junctions and hormone secretion influence conception rates in cows. ProcNatlAcadSci 110: 19495-19500.
- Sugimoto M, Gotoh Y, Kawahara T, Sugimoto Y (2015) Molecular effects of polymorphism in the 3’UTR of unc-5 homolog c associated with conception rate in holsteins. PLoS ONE 10: e0131283.
- Sugimoto M, Gotoh Y, Kawahara T, Sugimoto Y (2016) Family with sequence similarity 213, member A is associated with the fertility selection index in holsteins. PLoS ONE submitted.
- Kawahara T, Gotoh Y, Suzuki M, Baba T, Yamaguchi S (2012) Estimates of genetic parameters for female fertility traits and development of a fertility selection index in Japanese holsteins. Proceedings of 15th AAAP Animal Science Congress, Thammasat University, Rangsit Campus, Bangkok /PathumThani, Thailand, pp 26-30.
- Garrick DJ, Taylor JF, Fernando RL (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet SelEvol 41: 55.
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263-265.
- Markljung E, Jiang L, Jaffe JD, MikkelsenTS, Wallerman O, et al. (2009) ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoSBiol 7: e1000256.
- WittkoppPJ, HaerumBK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85-88.
- Sugimoto M, Watanabe T, Sugimoto Y (2012) The molecular effects of a polymorphism in the 5'UTR of solute carrier family 44, member 5 that is associated with birth weight in Holsteins. PLoS One 7: e41267.
- Holt RJ, Vandiedonck C, Willis-Owen SA, Knight JC, Cookson WO, et al. (2015) A functional AT/G polymorphism in the 5'-untranslated region of SETDB2 in the IgE locus on human chromosome 13q14. Genes Immun 16: 488-494.
- Sugimoto M, Sugimoto Y (2012) Variant in the 5' untranslated region of insulin-like growth factor 1 receptor is associated with susceptibility to mastitis in cattle. G3 (Bethesda) 2:1077-1084.
- Xu B, Washington AM, Hinton BT (2014) PTEN signaling through RAF1 proto-oncogene serine/threonine kinase (RAF1)/ERK in the epididymis is essential for male fertility. ProcNatlAcadSci 111: 18643-18648.
- Juma AR, Damdimopoulou PE, GrommenSV, Van de VenWJ, De Groef B (2016) Emerging role of PLAG1 as a regulator of growth and reproduction. J Endocrinol 228: R45-56.
- Kas K, Voz ML, Hensen K, Meyen E, Van de VenWJM (1998) Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J BiolChem 273: 23026-23032.
- Fortes MRS, Kemper K, Sasazaki S, Reverter A, Pryce JE, et al. (2013) Evidence for pleiotropism and recent selection in the PLAG1 region in Australian Beef cattle. Anim Genet. 44: 636-647.
- Hensen K, Braem C, Declercq J, Van Dyck F, DewerchinM, et al. (2004) Targeted disruption of the murine Plag1 proto-oncogene causes growth retardation and reduced fertility. Dev Growth Differ 46: 459-470.
- Gudbjartsson DF, Walters GB, ThorleifssonG, StefanssonH, Halldorsson BV, et al. (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40: 609-615.
- Cironi L, Riggi N, Provero P, Wolf N, Suvà ML, et al. (2008) IGF1 is a common target gene of Ewing's sarcoma fusion proteins in mesenchymal progenitor cells. PLoS One 3: e2634.
- RibeiroES, Bruno RG, Farias AM, Hernández-Rivera JA, Gomes GC, et al. (2014) Low doses of bovine somatotropin enhance conceptus development and fertility in lactating dairy cows. BiolReprod 90: 10.
- Ohta T, Koshi K, Ushizawa K, Hosoe M, Takahashi T, et al. (2014) Expression profiles of perforin, granzyme B and granulysin genes during the estrous cycle and gestation in the bovine endometrium. AnimSci J 85: 763-769.
- Hansen PJ, Soto P, Natzke RP (2004) Mastitis and fertility in cattle - possible involvement of inflammation or immune activation in embryonic mortality. Am J ReprodImmunol 51: 294-301.
- Bottazzi B, Bastone A, Doni A, Garlanda C, Valentino S, et al. (2006) The long pentraxin PTX3 as a link among innate immunity, inflammation, and female fertility. J LeukocBiol 79: 909-912.
- Minge CE, RobkerRL, Norman RJ (2008) PPAR Gamma: Coordinating Metabolic and Immune Contributions to Female Fertility. PPAR Res. 2008: 243791.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences