ISSN : 0976-8505
Der Chemica Sinica
Synthesis, Characterization, DNA Interaction and Anticancer Activity of Organotin(IV) Complexes with Sodium 3-(1H-indol-3-yl) propanoate
Shaheen F1, Ali S1*, Shahzadi S*1,2
1Department of Chemistry, Quaid-i-Azam University, Islamabad 45320, Pakistan
2Department of Chemistry, University of Sargodha, Lyallpur Campus, Faisalabad, Pakistan
Abstract
Six organotin(IV) complexes have been synthesized by refluxing sodium salt of 3-(1H-indol-3-yl) propanoic acid with di- and triorganotin chlorides in 1:1 and 2:1 molar ratios, respectively. These complexes have been characterized by elemental analysis, IR, 1H, 13C and 119Sn NMR spectroscopies. DNA binding studies were performed by viscometric measurements and UV-visible spectroscopy. Both techniques showed an intercalation mode of interaction. The cytotoxic activity of ligand salt and organotin(IV) complexes 1-6 was tested against human ovarian cell line A2780. Results of bioassay revealed that organotin derivatives were more active than anticancer drug cis-platin. Triorganotin(IV) compounds are found more potent for their cytotoxic activity as compared to corresponding diorganotin(IV) complexes.
Keywords
Organotin(IV) complexes, IR, NMR, DNA interaction, Cytotoxicity, Human ovarian cell line A2780
Introduction
Owing to intriguing variety of crystal structures and topologies, organotin(IV) carboxylates are thought to provide a better perceptive of biological systems, as there is a significant influence of the structure of the molecule and the coordination number of the tin atoms on the activities of organotin(IV) compounds [1,2].
It is known that the formation of metal carboxylates is strongly dependent on various factors including reaction conditions, such as pH values, temperature, solvent systems, stoichiometry, and choice of appropriate precursors as a source of carboxylate ligands [3]. Organotin(IV) complexes with carboxylates show immense structural variety. However, such a structural diversity makes it difficult in some cases to characterize them fully just by analytical techniques. Thus, X-ray crystallographic study makes full identification possible [4].
Organotin carboxylates find applications both in industry as PVC stabilizers, homogeneous catalysts, used as antifouling paints, flame retardant and smoke suppressants, antiwar agents, homogeneous catalysts and recycling agents as well as in agriculture as biocides due to their antibacterial and antifungal activities, as wood preservatives, biodiesel catalysts, antioxidants for polypropylene and anti-herpes agents [5-7].
Antitumor activities of some organotin(IV) carboxylates are reported in literature [8-12] and these are found to show promising cytotoxic activities against various cancer cells like, sarcoma cancer cells, lungs, liver, breast and colon carcinoma etc. Antitumor activities of some of the complexes are also found to be competing with cis-platin [9,12].
Extensive investigations regarding antitumor activity have been performed with organotin(IV) compounds, however the mechanism of their toxicity remains obscure. One of the assumptions may be that these compounds are capable of reacting with cell membranes leading to leakage, which accelerates ion exchange processes, and inhibits oxidative or photochemical phosphorylation [13]. We have synthesized organotin(IV) complexes with sodium salt of 3-(1H-indol- 3-yl) propanoic acid and characterized by IR, multinuclear (1H, 13C and 119Sn) NMR. DNA binding studies and anticancer activity were also performed.
Materials and Methods
Trimethyltin chloride (97%), tributyltin chloride (96%), triphenyltin chloride (95%), dimethyltin chloride (97%), dibutyltin chloride (96%) and diphenyltin dichloride (96%) used were purchased from Sigma Aldrich. Analytical grade solvents were purchased from E. Merck and Fluka and were dried by standard procedures [14]. Melting points were determined in capillary tube using electrothermal melting point apparatus model MP-D Mitamura Riken Kogyo (Japan) and were uncorrected. Elemental analysis was done using a thermoscientific Flash 2000 CHN/S organic elemental analyser. Infrared spectra were recorded as KBr/CsBr discs on Perkin Elmer spectrum 1000 (USA) in range of 4000–250 cm-1. 1H and 13C NMR were recorded on Bruker AC 300 MHFT-NMR. 119Sn NMR spectra were taken on Avance 400 MHz TXO 10 mm XWINNMR. Me4Sn was used as standard reference. Viscosity was measured by an Ubbelohde viscometer at room temperature. The absorption spectra were recorded on a Shimadzu 1800 UV–Vis spectrophotometer.
Synthesis
Synthesis of sodium 3-(1H-indol-3-yl) propanoate NaL
Sodium salt of 3-(1H-indol-3-yl) propanic acid was prepared by 1:1 molar reaction of 3-(1H-indol-3-yl) propanoic acid with aqueous solution of sodium hydroxide in methanol with continuous stirring at room temperature for 1 hr. The product obtained was soluble in methanol and was isolated by evaporating the solvent under vacuum. Ligand salt thus obtained was washed with diethyl ether and was air dried.
(Yield: 96.05 %). M.p: 277-279 °C. FT-IR (cm-1): νas (COO) 1618, νs (COO) 1387, Δν (231). 1H-NMR (ppm): 1.94 (t, H2, 3J=7 Hz), 2.14 (t, H3, 3J=7Hz), 10.65 (N-H), 6.87 (s, H12), 7.02-7.36 (m, Ar-H6-9). 13C-NMR (ppm):203.1 (C-1), 42.1 (C-2), 39.8 (C-3), 108.3 (C-4), 121.5 (C-12), 137.1, 125.4, 127.8, 128.8, 130.8 134.2 (Ar-C).
General procedure for synthesis of complexes 1-6
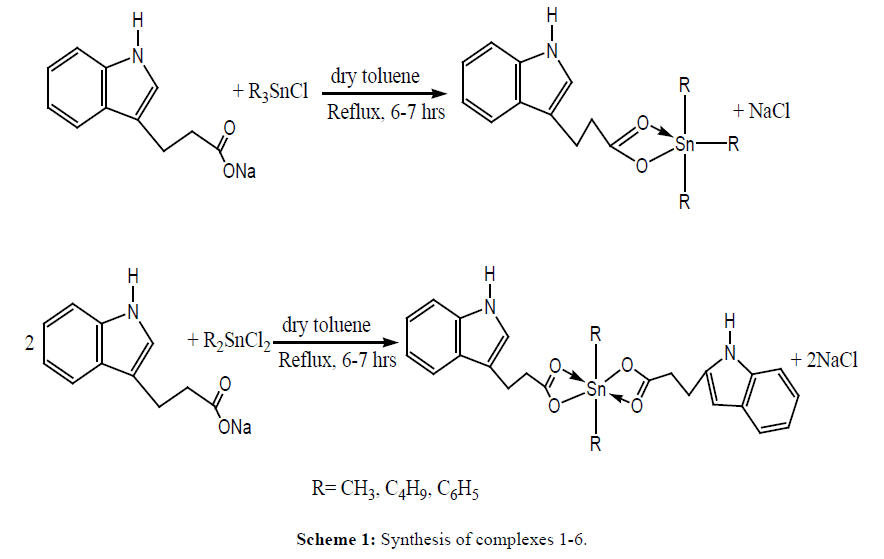
Tri and diorganotin(IV) complexes were synthesized by refluxing ligand salt with tri and diorganotin chlorides in 1:1 and 2:1 molar ratios, respectively in dry toluene for 6-7 hrs. The NaCl formed was removed by filtration and solvent was evaporated under vacuum. All synthesized compounds were recrystallized in chloroform: n-hexane mixture (4:1) (Scheme 1).
Trimethylstannyl 3-(1H-indol-3-yl)propanoate 1: (Yield: 71.4 %). M.p:194°C, Elemental analysis, % Calculated (Found), for C14H19NO2Sn: C, 47.77 (47.82); H, 5.44 (5.49); N, 3.98 (4.02). FT-IR (cm-1): νas (COO) 1575, νs (COO) 1395, Δν (180). 1H-NMR (ppm): 2.13(t, H2, 3J=7Hz), 2.45 (t, H3, 3J=7Hz), 10.63 (N-H), 6.87 (s, H12), 7.04-7.35 (m, Ar-H6-9), 0.89(s, CH3-Sn, [56]).13C-NMR (ppm): 198.6 (C-1), 42.3 (C-2), 39.8 (C-3), 108.5 (C-4), 123.2 (C-12) 135.1, 125.8, 126.0, 128.8, 127.8 134.2 (Ar-C), 2.6 {(CH3-Sn), 1J[119Sn-13C=352 Hz]}.119Sn NMR (CDCl3, ppm): -10
Tributylstannyl 3-(1H-indol-3-yl)propanoate 2: (Yield: 75.22 %). Elemental analysis, % Calculated (Found), for C23H37NO2Sn: C, 57.76 (57.80); H, 7.80 (7.93); N, 2.93 (2.84). FT-IR (cm-1): νas (COO) 1583, νs (COO) 1396, Δν (187). 1H-NMR (ppm): 2.13 (t, H2, 3J=7Hz), 2.46 (t, H3, 3J=7Hz), 10.54 (N-H), 6.86 (s, H12), 7.05-7.38 (m, Ar-H6-9), 1.21-1.63 (m, H- α, H- β,H- γ ), 0.87 (t, Hδ, 3J=7.4Hz ).13C-NMR (ppm): 197.9 (C-1), 41.8 (C-2), 39.5 (C-3), 108.3 (C-4), 122.2 (C-12) 135.4, 125.7, 126.1, 128.8, 127.6 134.4 (Ar-C), 13.4 {(C- α,1J[119Sn-13C=356 Hz]}, 23.2 (C- β ), 25.5 (C- γ ), 28.3 (C- δ ). 119Sn NMR (CDCl3, ppm): -29
Triphenylstannyl 3-(1H-indol-3-yl)propanoate 3: (Yield: 73.53 %). M.p: 102-104°C, Elemental analysis, % Calculated (Found), for C29H25NO2Sn: C, 64.71 (64.75); H, 4.68 (4.64); N, 2.60 (2.69). FT-IR νas (COO) 1594, νs (COO) 1406, Δν (188).1H-NMR (ppm): 2.14 (t, H2, 3J=7Hz), 2.45 (t, H3, 3J=7Hz), 10.59 (N-H), 6.85 (s, H12), 7.06-7.38 (m, Ar-H6-9), 7.43-7.52 (m, Sn-C6H5),7.49-7.85 (m, Sn-C6H5).13C-NMR (ppm):197.1 (C-1), 42.2 (C-2), 39.5 (C-3), 108.1 (C-4), 122.5 (C-12) 134.5, 125.4, 126.5, 128.8, 127.1 134.0 (Ar-C),139.5 (C- α ), 136.9(C- β ), 129.1 (C- γ ), 131.2 (C- δ ). 119Sn NMR (CDCl3, ppm): -221
Dimethylstannanediyl bis[3-(1H-indol-3-yl)propanoate 4: (Yield: 64.11 %). M.p: 150-156°C, Elemental analysis, % Calculated (Found), for C24H26N2O4Sn: C, 54.89(54.65); H, 4.99 (4.94); N, 5.33 (5.27). FT-IR (cm-1): νas (COO) 1589, νs (COO) 1408, Δν (181). 1H-NMR (ppm): 2.15 (t, H2,2’, 3J=7Hz), 2.45 (t, H3,3’, 3J=7Hz), 10.62 (N-H), 6.82(s, H12,12,), 7.09-7.42 (m, Ar-H6-9), 0.95 (s, CH3-Sn [60]). 13C-NMR (ppm): 197.0 (C-1), 42.3 (C-2), 39.8 (C-3), 108.7 (C-4), 122.5 (C-12) 135.5, 125.2 126.9, 128.8, 127.5 134.1 (Ar-C), 1.4 {(CH3-Sn), 1J[119Sn-13C=352 Hz]}. 119Sn NMR (CDCl3, ppm): -45
Dibutylstannanediyl bis[3-(1H-indol-3-yl)propanoate 5: (Yield: 71.74 %). M.p: 94-98 °C, Elemental analysis, % Calculated (Found), for C30H38N2O4Sn: C, 59.13 (59.17); H, 6.29 (6.24); N, 4.60 (4.65). FT-IR (cm-1): νas (COO) 1582, νs (COO) 1403, Δν (179). 1H-NMR (ppm):2.13 (t, H2,2’, 3J=7Hz), 2.44 (t, H3,3’, 3J=7HZ), 10.62 (N-H), 6.86 (s, H12,12’), 7.05-7.36 (m, Ar-H6-9),1.25-1.59 (m, H- α, H- β, H- γ, H- δ ).13C-NMR (ppm): 197.3 (C-1), 42.2 (C-2), 39.9 (C-3), 108.4 (C-4), 122.5 (C-12) 135.8, 124.9, 126.7, 129.0, 127.5 134.7 (Ar-C),12.9, 17.3, 26.5, 29.3 (C- α, C- β, C- γ, C- δ ).119Sn NMR (CDCl3, ppm): 65
Diphenylstannanediyl bis[3-(1H-indol-3-yl)propanoate 6: (Yield: 70.32 %). M.p: 128-131 °C, Elemental analysis, % Calculated (Found), for C34H30N2O4Sn: C, 62.89 (62.84); H, 4.66 (4.70); N, 4.31 (4.35). FT-IR (cm-1): νas (COO) 1587, νs (COO) 1392, Δν (195). 1H-NMR (ppm): 2.14 (t, H2,2’,3J=7Hz), 2.45 (t, H3,3’, 3J=7Hz), 10.58 (N-H), 6.85 (s, H12,12’), 7.06-7.38 (m, Ar-H6-9), 7.43-7.52 (m, 6H), 7.81-7.85 (m, 4H).13C-NMR (ppm): 197.1 (C-1), 42.3 (C-2), 39.1 (C-3), 108.9 (C-4), 122.4 (C-12) 135.5, 125.5, 126.3, 128.7, 127.3 134.1 (Ar-C), 139.2 (C- α ), 137.0 (C- β ), 128.3 (C- γ ), 133.2 (C- δ ), 119Sn NMR (CDCl3, ppm): -197
Cytotoxic activity
Cytotoxic potential of compounds 1-6 was performed by MTT-Method [15], to evaluate the effect of compounds on the growth of cell. The human ovarian carcinoma cell line (A2780) was chosen for this purpose. This method is based on principle of reduction of yellow colored tetrazolium salt, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] to purple formazan product, by the mitochondrial dehydrogenase of metabolically active cells. MTT solution was prepared by dissolving it at concentration of 5 mg/ml in phosphate buffer saline. Then 20 μL of this solution was added to each of 96 well plates containing 100 μL of culture medium and was incubated at 37ºC for 3 to 4 hr. The medium was aspirated carefully without disruption of purple coloured formazan crystals. The resulting purple colour zone was solubilized in 100 μL of DMSO and quantified by spectrophotometer. Micro ELISA plate reader was used to read the plate at wavelength of 590 nm to determine IC50 value.
Results and Discussion
FT-IR Spectroscopy
FT-IR spectra of ligand and compounds 1-6 were recorded in the range of 4000-250 cm-1. Asymmetric and symmetric stretching vibrations (-COO) are very useful in describing structure of organotin(IV) carboxylates. The νasym (COO) stretch appeared in the range of 1575-1618 cm-1, while vibrations observed in region of 1408-1387 cm-1 are characteristic of νsym (COO). It is reported in literature [16] that difference Δν (COO) above 200 cm-1 suggests a monodentate coordination, while difference below 200 cm-1 propose a bidentate coordination. In all synthesized compounds, the Δν value comes out to be less than 200 cm-1 which suggest a chelating or bridging mode of coordination [17,18], therefore, tri- and diorganotin(IV) complexes exhibits five and six coordinated geometry, respectively in solid state.
Multinuclear NMR Spectroscopy
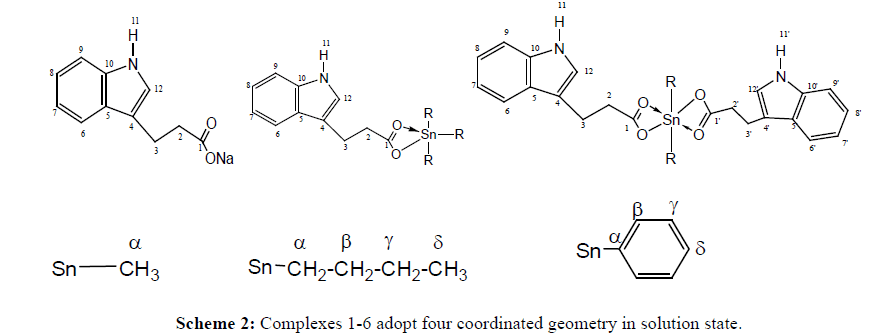
1H, 13C and 119Sn NMR of compounds 1-6 were recorded in deuterated chloroform. In 1H and 13C NMR spectra new signals for alkyl and aryl groups in corresponding region confirms complexation. In 1H NMR, 2J[119Sn-1H] coupling constant values obtained for complexes 1 and 4 depicted a tetrahedral geometry in solution which is in accordance with 1J[119Sn-13C]. Also 1J [119Sn-13C] coupling constant obtained for compound 2 confirms a tetrahedral environment around tin. Unfortunately, couplings were not observed for phenyl complexes in either 1H or 13C NMR due to complex pattern; however ipso carbon gave a signal at 139.5 and 139.2 ppm for compound 3 and 6, respectively. N-H proton of indole unit appeared as a singlet in all complexes which propose that N atom of indolyl unit does not coordinate to Sn centre. It is apparent from these observations that complexes 1-6 adopt four coordinated geometry in solution state (NMR numbering is given as Scheme 2).
To further confirm solution state geometry, 119Sn NMR spectra of complexes were recorded in deuterated chloroform.119Sn chemical shift of organotin(IV) compounds cover a range of ± 600 ppm [19]. It is reported that in alkyltin carboxylates, tetra-coordinated tin has δ (119Sn) values ranging from about +200 to -60 ppm, and pentacoordinated tin from -200 to -400 ppm [20]. A sharp signal appearing at -10, -29, -45, and 65 ppm indicates a 4-coordinated geometry in solution for compounds 1, 2, 4 and 5, respectively. For triphenyltin complexes, chemical shift in the range of -180 to -260 is an indicative of tetrahedral geometry [21]. In present case δ value of -221 and -197 ppm for complexes 3 and 6, respectively confirms a tetrahedral configuration in solution. Thus, 119Sn NMR results are in good congruence with 1H and 13C NMR results.
Evaluation of DNA binding parameters
DNA binding studies of sodium 3-(1H-indol-3-yl) propanoate (L-salt) and its organotin(IV) derivatives (complexes 1, 2 and 3) were carried out by using UV-vis spectroscopy and viscometry. Solutions of ligand salt and metal complexes were prepared in DMSO.
Viscosity measurement
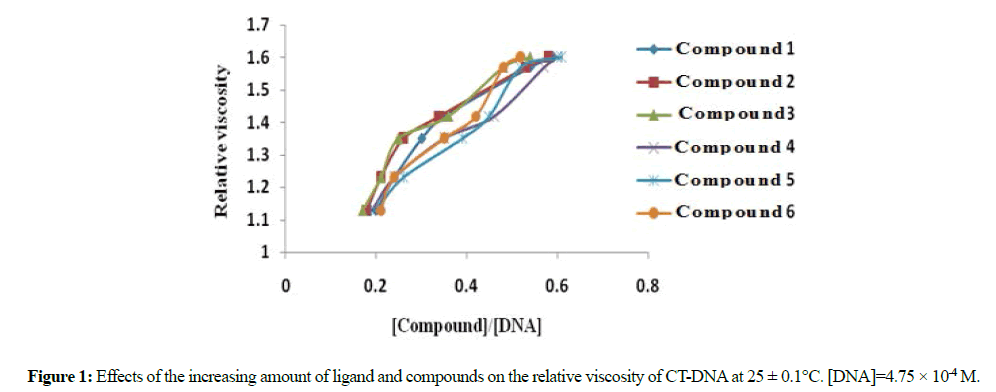
The viscosity of DNA is sensitive to its length change. Mode of DNA binding to a compound can be well assigned by relative viscosity measurements which have been proved to be most reliable and least ambiguous technique. In the case of classic intercalation, DNA base pairs are separated in order to accommodate the bound compound, resulting in the lengthening of the DNA helix and consequently increased DNA viscosity [22-26]. On the other hand, the binding of a compound exclusively in DNA grooves by means of partial and/or non-classic intercalation, under same conditions, (e.g., netropsin, distamycin), causes a bend or kink in the DNA helix, reducing its effective length and, as a result, the DNA solutions viscosity is decreased or remains unchanged, i.e. groove binders and electrostatic interaction do not lengthen the DNA molecules [27-29].
Different sets of solution were prepared with a fixed DNA concentration (4.75 × 10-4 M) and varying the concentration of ligand salt and complexes. The values of the relative viscosity (η/η₀)1/3 were calculated from (t/to)1/3 ratio for all samples, where t, to, η and η₀ represent the time and viscosity of the DNA solution with and without compound, respectively. A plot of (η/η₀)1/3 versus the [compound]/[DNA] ratio shows that the viscosity increases with increasing concentration of the compounds, which suggests an intercalation mode of binding for ligand and all compounds.
Electronic absorption spectra
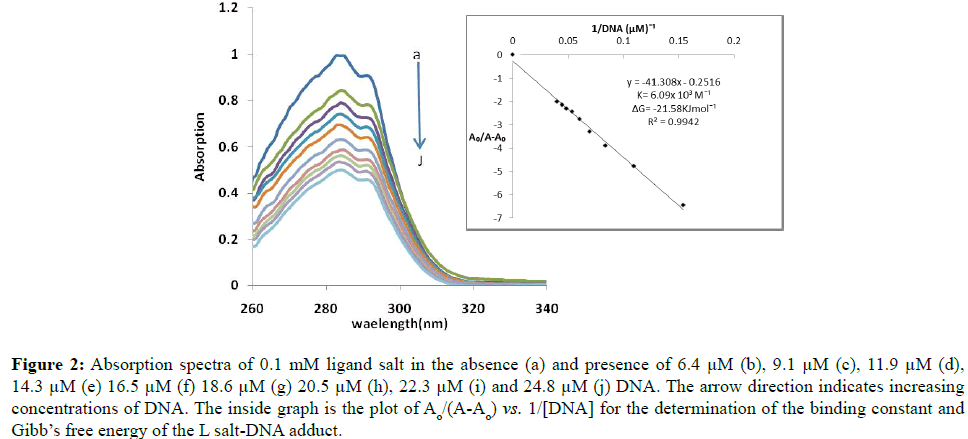
The absorption spectra of ligand salt and compounds 1, 2 and 3 in the absence and presence of different DNA concentrations are given as Figures 1-5. Two transitions bands were observed due to π - π* transitions. In all the compounds, both peaks showed hypochromism which suggest the intercalative mode of interaction between complexes and DNA. Decrease in transition probability as a consequence of interaction of partly filled π* orbital of ligand with π orbital of DNA base pairs may be the possible reason of hypochromism. There is only slight red shift of 1-2 nm, thus lack of any appreciable red shift in all compounds suggests the weak interactions with DNA [30].
Figure 2: Absorption spectra of 0.1 mM ligand salt in the absence (a) and presence of 6.4 μM (b), 9.1 μM (c), 11.9 μM (d), 14.3 μM (e) 16.5 μM (f) 18.6 μM (g) 20.5 μM (h), 22.3 μM (i) and 24.8 μM (j) DNA. The arrow direction indicates increasing concentrations of DNA. The inside graph is the plot of Ao/(A-Ao) vs. 1/[DNA] for the determination of the binding constant and Gibb’s free energy of the L salt-DNA adduct.
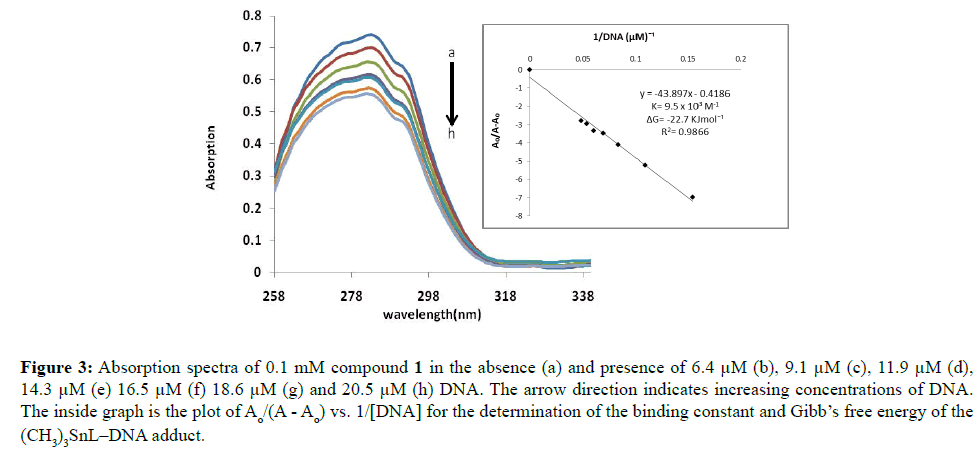
Figure 3: Absorption spectra of 0.1 mM compound 1 in the absence (a) and presence of 6.4 μM (b), 9.1 μM (c), 11.9 μM (d), 14.3 μM (e) 16.5 μM (f) 18.6 μM (g) and 20.5 μM (h) DNA. The arrow direction indicates increasing concentrations of DNA. The inside graph is the plot of Ao/(A - Ao) vs. 1/[DNA] for the determination of the binding constant and Gibb’s free energy of the (CH3)3SnL–DNA adduct.
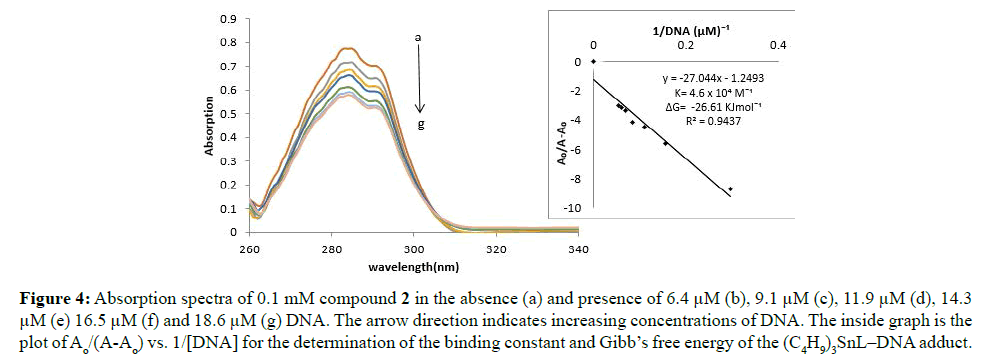
Figure 4: Absorption spectra of 0.1 mM compound 2 in the absence (a) and presence of 6.4 μM (b), 9.1 μM (c), 11.9 μM (d), 14.3 μM (e) 16.5 μM (f) and 18.6 μM (g) DNA. The arrow direction indicates increasing concentrations of DNA. The inside graph is the plot of Ao/(A-Ao) vs. 1/[DNA] for the determination of the binding constant and Gibb’s free energy of the (C4H9)3SnL–DNA adduct.
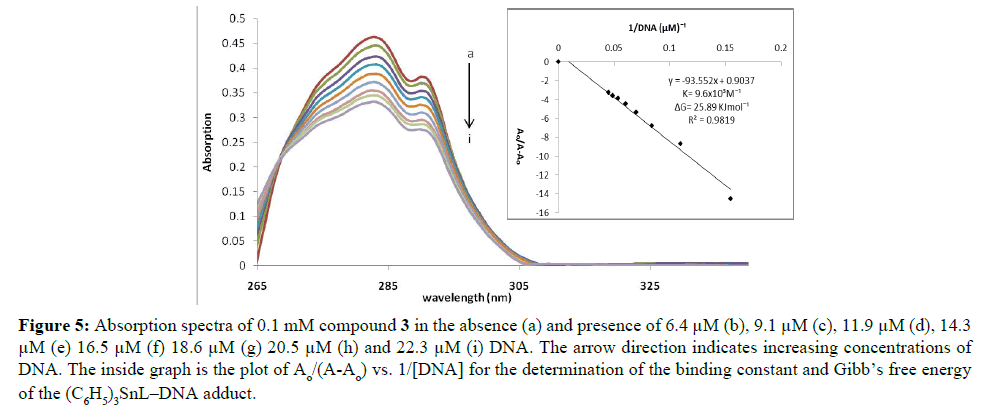
Figure 5: Absorption spectra of 0.1 mM compound 3 in the absence (a) and presence of 6.4 μM (b), 9.1 μM (c), 11.9 μM (d), 14.3 μM (e) 16.5 μM (f) 18.6 μM (g) 20.5 μM (h) and 22.3 μM (i) DNA. The arrow direction indicates increasing concentrations of DNA. The inside graph is the plot of Ao/(A-Ao) vs. 1/[DNA] for the determination of the binding constant and Gibb’s free energy of the (C6H5)3SnL–DNA adduct.
Benesi-Hildebrand equation [19] was used to calculate binding constant of these complexes.
A0/(A-A0)=ÆÂÂG/(ÆÂÂH-G-ÆÂÂG)+ ÆÂÂG/(ÆÂÂH-G-ÆÂÂG) × 1/K[DNA] (1)
In equation 1, K is the association constant, A0 and A are the absorbance of the compound and its complex with DNA, respectively, while ÆÂÂG and ÆÂÂH-G are the absorption coefficients of the compound and the compound-DNA complex, respectively.
The binding constants were calculated from the intercept-to-slope ratios of straight line graphs obtained from A0/ (A-A0) vs. 1/[DNA] plots. A comparison in binding constants suggest that compound 2 forms most stable complex with DNA as compared to ligand salt, complexes 1 and 3. Gibb’s free energy (ΔG) was calculated by using following equation [31].
ΔG= -RTlnK
Where, R=8.314 JK-1mol-1(general gas constant), T=298 K (room temperature). Negative value of ΔG in all compounds showed that compound–DNA adduct formation is a spontaneous process.
Cytotoxic activity
Cellular viability of tumor cell line A2780 was tested with the ligand and complexes 1-6, by using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay [15].
Cis-platin was used as a reference drug. The results of cytotoxic activities were determined as IC50 which corresponds to the minimum concentration of compounds in μgmL-1 that inhibits proliferation rate of the tumor cells by 50% in comparison to control untreated cells. All the tri- and diorganotin(IV) compounds showed good cytotoxic activities as compared to cis-platin and reported organotin compounds [32-34] and exhibited lower IC50 values (Table 1). The biological activity of organotin complexes is influenced by the nature of the ligand environment, organic groups attached to the tin, compound structure, toxicity, and potential mechanism of action. The function of the ligand is to support the transport of the active organotin moiety to the site of the action where it is released by hydrolysis [35]. The anionic ligand also plays an important role in determining the degree of the activity of organotin compounds. As all complexes showed tetrahedral geometry in solution and showed significant activity, our results are consistent with the literature that states species generating a tetrahedral geometry in solution are more active [36]. The coordination number of the triorganotin derivatives are usually four in non-coordinating solvents while for diorganotin derivatives it vary from 4 to 6 due to fluxional behaviour/size of the ligands attached. As for as DMSO is concerned, it is a coordinating solvent and it might be possible that DMSO could change the coordination number of the complex to 5 or 6. However, in biological system when the complex reaches the target site having sulphur or nitrogen as donor for coordination, DMSO can vacate the site as a weak/hard donor [37,38].
| Compound | IC50 |
|---|---|
| 1 | 4.5 ± 0.1 |
| 2 | 4.0 ± 1.0 |
| 3 | 4.3 ± 0.9 |
| 4 | 7.2 ± 0.7 |
| 5 | 6.2 ± 0.4 |
| 6 | 8.2 ± 0.2 |
| Cisplatin | 7.5 ± 0.2 |
Table 1: IC50 values (μg/ml) of compounds 1-6 tested against human ovarian tumor cell lines A2780a.
Activity and selectivity of complexes may also be related to the finding of recently reported organotin(IV) complexes against various other cell lines (i.e. Human lung tumorA549, hepatocellular liver carcinom HepG2, human colon cancer HCT 116, breast cancer MCF-7, leukemia K562 and normal cell line (3T3-L1) [39-41]. Additionally orientational and substitutional effects may also be considered in this connection.
Conclusion
It has been concluded that sodium salt of 3-(1H-indol-3-yl) propanoic acid acts as bidentate and complexes exhibited 5 or 6 coordinated geometry in solid state. While multinuclear NMR (1H,13C,119Sn) confirmed the tetrahedral geometry in solution. The complexes bind effectively with DNA and possessed good antitumor activity against ovarian carcinoma, at a concentration of 0.1 mM, thereby increasing their therapeutic value. Organotin(IV) complexes were found more active as compared to cis-platin, while triorganotin(IV) complexes are shown to possess greater cytotoxicity in comparison to diorganotin(IV) compounds.
References
- Yin HD, Gao ZJ, Wang CH (2005) Synthesis, characterization and properties of organotin complexes dibutyltin(IV) bis(heteroaromatic carboxylate) and crystal structure of dibutyltin(IV) bis(2-thiazolylcarboxylate). Chin J Chem 23: 928-932.
- Win YF, Teoh SG, Vikneswaran TMR, Chan MY, Ha ST et al. (2010) Synthesis and characterization of organotin(IV) complexes derived of 3-(dimethylamino)benzoic acid: Cytotoxic assay on human liver carcinoma cells (HepG2). Am J Appl Sci 7: 301-308.
- Tao B, Xia H, Huang C-X, Li X-W (2011) Hydrolytic ring opening reactions of pyromellitic dianhydride for divalent transition metal carboxylate complexes. Anorg Z Allg Chem 631: 703-707.
- Teoh SG, Ang SH, Looi ES, Keok CA, Teo SB et al (1997) Novel hydroxo bidentate carboxylate bridging in diorganotin(IV) carboxylate complex Synthesis and crystal structure of μ-hydroxo-μ-trichloroacetato-O,O′-bis (trichloroacetato-O)-bis[dibutyltin(IV)], Sn2(C4H9)4(O2CCCl3)3(OH). J Organomet Chem 527: 15-19.
- Shahzadi S, Shahid K, Ali S (2007) Coordination behaviour based on spectroscopic studies of carboxylate group in organotin(IV) derivatives of 2-[(2`,4`,6`- tribromophenylamido)]benzoic acid and [(2`,4`,6`-tribromophenylamido)]propenoic acid. Russ J Coord Chem 33: 403-411.
- Bonire JJ, Ayoko GA, Olurinola PF, Ehinmidu JO, Jalil NSN et al (1998) Synthesis and antifungal activity of some organotin(iv) carboxylates. Met Based Drugs 5: 233-236.
- Tariq M, Ali S, Khalid N (2012) Activity of homogeneous and heterogeneous catalysts, spectroscopic and chromatographic characterization of biodiesel:A review. Renew Sust Ener Rev 16: 6303-6316.
- Balas VI, Hadjikakou SK, Hadjiliadis N, Kourkoumelis N, Light ME et al(2008) crystal structure and antitumor activity of the novel zwitterionic complex of tri-butyltin(IV) with 2-thiobarbituric acid. Bioinorg Chem & Appl 1: 1-5.
- Gielen M (1995) Tin-based antitumour drugs: New developments. Met Based Drugs 2: 99-103.
- Li W, Zhang ZW, Ren SM, Sibiril Y, Massin DP et al(2009) synthesis and antitumor activity of novel dibutyltin carboxylates of aminoglucosyl derivatives. Chem Biol Drug Des 73: 682-686.
- Kang W, Wub X, Huang J (2009) Synthesis, crystal structure and biological activities of four novel tetranuclear di-organotin(IV) carboxylates. J Organomet Chem 694: 2402-2408.
- Danish M, Ali S, Shahid K, Mazhar M (2004) Synthesis and in vitro antitumour studies of trimethyltin(IV) trans-m-methylcinnamate. J Chem Soc Pak 26: 140-142.
- Ahmad MS, Mirza B, Hussain M, Hanif M, Ali S et al(2008) ATR-FTIR spectroscopy detects alterations induced by organotin(IV) carboxylates in MCF-7 cells at sub-cytotoxic/-genotoxic concentrations. PMC Biophys 1: 1-19.
- Armarego WFL, Chai C (2003) Purification of laboratory chemicals, 5th ed., Butterworth, Oxford, USA.
- Mosman T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Immunol J Methods 65: 55-63.
- Hussain H, Ahmad VU, Green IR, Krohn K, Hussain J et al (2007) Antibacterial organotin(IV) compounds, their synthesis and spectral characterization. Arkivoc 14: 289-299.
- Abdel-Rahman LH, Abu-Dief AM, Adam MSS, Hamdan, SK (2016) Some new mono sized mononuclear Cu(II) schiff base complexes: Design, characterization, molecular modeling and catalytic potentials in benzoyl alcohol oxidation. Catal. Lett 146: 1373-1396.
- Abdel-Rahman LH, Abu-Dief AM, Mostafa H, Hamdan SK (2017) Sonochemical synthesis, structural inspection and semiconductor behavior of three new nano sized Cu(II), Co(II) and Ni(II) chelates based on tri-dentate NOO imine ligand as precursors for preparing of metal oxides. Appl Organomet Chem 31: e3555.
- Wrackmeyer B (1999) Application of 119Sn NMR parameters. Annu Rep NMR Spectrosc 38: 203-264.
- Ahmad MS, Hussain M, Hanif M, Ali S, Mirza B (2007) Synthesis, chemical characterization and biological screening for cytotoxicity and antitumor activity of organotin (IV) derivatives of 3,4-methylenedioxy 6-nitrophenylpropenoic acid. Molecules 12: 2348-2363
- Sadiq-ur-Rehman, Ali S, Shahzadi S (2008) Organotin(IV) esters of (e)-3-furanyl-2-phenyl-2-propenoic acid: Synthesis,investigation of the coordination modes by IR, multinuclear NMR (1H, 13C, 119Sn) and in vitro biological studies. Heteroatom Chem 19: 612-620.
- Abdel-Rahman LH, El-Khatib RM, Nassr LAE, Abu-Dief AM, Lashin FE (2013) Design, characterization, teratogenicity testing, antibacterial, antifungal and DNA interaction of few high spin Fe (II) Schiff base amino acid complexes. Spectrochim Acta A 111: 266-276.
- Abdel-Rahman LH, El-Khatib RM, Nassr LAE, Abu-Dief AM, Ismael M et al (2014) Metal based pharmacologically active agents: synthesis, structural characterization, molecular modeling, CT-DNA binding studies and in vitro antimicrobial screening of iron(II) bromosalicylidene amino acid chelates. Spectrochim Acta 117: 366-378.
- Abu-Dief AM, Nassr LAE (2015) Tailoring, physicochemical characterization, antibacterial and DNA binding mode studies of Cu(II) schiff bases amino acid bioactive agents incorporating 5-bromo-2-hydroxybenzaldehyde. J Iran Chem Soc 12: 943-955.
- Abdel-Rahman LH, Abu-Dief A.M, Ismael M, Mohamed MAA, Hashem NA (2016) Synthesis, structure elucidation, biological screening, molecular modeling and DNA binding of some Cu(II) chelates incorporating imines derived from amino acids. J Mol Stru 1103: 232–244.
- Abdel-Rahman LH, Ismail NM, Ismael M, Abu-Dief AM, EA Ahmed (2017) Synthesis, characterization, DFTcalculations and biological studies of Mn(II), Fe(II), Co(II) and Cd(II) complexes based on a tetradentate ONNO donor Schiff base ligand. J Mol Struct 1134: 851-862.
- Abdel-Rahman LH, Abu-Dief AM, Hamdan SK, Seleem AA (2015) Nano structure iron (ii) and copper (ii) schiff base complexes of a NNO-tridentate ligand as new antibiotic agents: spectral, thermal behaviors and DNA binding ability. Int J Nano Chem 1 (2): 65-77.
- Abdel-Rahman LH, Abu-Dief AM, Newair EF, Hamdan SK (2016) Some new nano-sized Cr(III), Fe(II), Co(II), and Ni(II) complexes incorporating 2-((E)-(pyridine-2-ylimino)methyl)napthalen-1-ol ligand: Structural characterization, electrochemical, antioxidant, antimicrobial, antiviral assessment and DNA interaction. J Photochem Photobiol B 160: 18-31.
- Abdel-Rahman LH, Abu-Dief AM, Aboelez MO, Abdel-Mawgoud AAH (2017) DNA interaction, antimicrobial, anticancer activities and molecular docking study of some new VO (II), Cr (III), Mn (II) and Ni (II) mononuclear chelates encompassing quaridentate imine ligand. J Photochem Photobiol B 170: 271-285.
- Sirajuddin M, Ali S, Haider A, Shah NA, Shah A et al (2012) Synthesis, characterization, biological screenings and interaction with calf thymus DNA as well as electrochemical studies of adducts formed by azomethine [2-((3,5-dimethylphenylimino)methyl)phenol] and organotin(IV) chlorides. Polyhedron 40: 19-31.
- Kalanur SS, Katrahalli U, Seetharamappa J (2009) Electrochemical studies and spectroscopic investigations on the interaction of an anticancer drug with DNA and their analytical applications. J Electroanal Chem 636: 93-100.
- Shahzadi S, Shahid K, Ali S, Malik A, Ahmed E (2005) Synthesis, spectroscopic studies and antileishmal, antibacterial, antifungal, cytotoxicity and insecticidal activities of organotin(iv) derivatives of (e)-4-oxo-4-(2,4,6-trichloroanilino)-2-butenoic acid. J Chem Soc Pak 27: 541-547.
- Shahid K, Shahzadi S, Ali S, Mazhar M (2006) Synthesis, spectroscopic studies and biological applications of organotin(iv) derivatives of 3-[n-(4-nitrophenyl)-amido]propenoic acid and 3-[n-(4-nitrophenyl)-amido]propanoic acid. Bull Korean Chem Soc 27: 44-52.
- Shahzadi S, Shahid K, Ali S (2007) Coordination behavior of the carboxylate group in organotin(iv) derivatives of 2-[(2',4',6'-tribromophenylamido)]benzoic acid and 3-[(2',4',6'-tribromophenyl-amido)] propenoic acid: Spectroscopic studies. Russian. J Coord Chem 33: 403-411.
- Shahzadi S, Ali S, Fettouhi M (2008) Synthesis, spectroscopy, in vitro biological activity and x-ray structure of (4-methylpiperidine-dithiocarbamato-s,s’)triphenyltin(iv). J Chem Cryst 38: 273-278.
- Molloy KC, in: Hartley FE, (Ed.) (1989) Bioorganotin compounds, the chemistry of metal- carbon bond. Wiley, New York, USA.
- Danish M, Alt HG, Badshah A, Ali S, Mazhar M et al (1995) Organotin esters of 3-(2-furanyl)-2-propenoic acid: Their characterization and biological activity. J Organomet Chem 486: 51-56.
- Danish M, Ali S, Mazhar M (1996) Fluxional behaviour in dimeric tetraorganodicarboxylatostannoxanes. Heteroatom Chem 7: 233-237.
- Zhang RF, Yan PZ, Li QL, MaCL (2014) Novel μ2-oxo-bridged dinuclear aryltelluronic triorganotin(IV) esters: syntheses, structural characterizations, and in vitro cytotoxicity. J Coord Chem 67: 649-659.
- Win YF, Choong CS, Dang JC, Iqbal MA, Quah CK et al (2014) Polymeric seven-coordinated organotin(IV) complexes derived from 5-amino-2-chlorobenzoic acid and in vitro anti-cancer studies. J Coord Chem 67: 3401-3413.
- Haque RA, Salam MA, Arafath MdA (2015) New organotin(IV) complexes with N(4)-methylthiosemicarbazone derivatives prepared from 2,3-dihydroxybenzaldehyde and 2-hydroxy-5-methylbenzaldehyde: synthesis, characterization, and cytotoxic activity. J Coord Chem 68: 2953-2967.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences