Molecular Characteristics of Probiotics Lactic Acid Bacteria Isolated from Soursop, Cowmilk, Goatmilk Yoghurts and Cheese
Department of Food Technology, University of Ibadan, Nigeria
- *Corresponding Author:
- Abiona SO
Department of Food Technology
University of Ibadan, Nigeria
Tel: +2349033296181
E-mail: abbsoy111@yahoo.com
Received Date: November 29, 2017; Accepted Date: December 14, 2017; Published Date: December 21, 2017
Citation: Abiona SO, Adegoke GO (2017) Molecular Characteristics of Probiotics Lactic Acid Bacteria Isolated from Soursop, Cowmilk, Goatmilk Yoghurts and Cheese. J Food Biotechnol Res. Vol.1 No.1:7
Abstract
Probiotics are living, non-pathogenic microorganisms which has positive effect on consumers. Yoghurt is among the most common dairy products consumed around the world. Different varieties of wild fruits could be of health benefit, one such is soursop fruit (Annona muricata). They are fruits that are good sources of vitamins and minerals. Goat milk is naturally predisposed for preparing a yoghurt drink that is light and easily digestible. Fermented drinks made of goat milk differ from the drinks made of cow milk, among other things, by a lower content of volatile aroma compounds (and especially of acetaldehyde and carbon dioxide. Cheese is white soft non-ripened milk made by the addition of a plant extract (Calotropis procera) to the non-pasteurized whole milk from cattle. The study was aimed at isolation and molecular characterization of lactic acid bacteria (LAB) from soursop, cowmilk, goatmilk yoghurts and cheese. Twenty-four bacterial strains were isolated, according to biochemical identification and only 16 were characterize as: Lactobacillus plantarium, Lactobacillus fermentus, Bacillus substilis, Bacillus subsitils, Lactobacillus rhaminosus, Lactobacillus fermentum, Lactobacillus paracase, Lactobacillus licheniformis, Lactobacillus planetarium, Lactobacillus licheniformis, Bacillus substilis, Lactobacillus paracasei, Lactobacillus fermentus, Lactobacillus fermentus, Lactobacillus licheniformis, Lactobacillus rhaminouus, Lactobacillus licheniformis. The isolates were characterize on the basis of their morphological, biochemical, physiological and 16S rRNA gene sequences. All of the isolates were well identified with the help of molecular techniques. The study concluded that the isolated lactic acid bacteria could be considered as potential candidates for starter culture in dairy industry.
Keywords
Yoghurt; Lactic acid bacteria; Molecular characterization
Introduction
Lactic acid bacteria (LAB) are widely used in fermented food production and are considered as generally recognized as safe (GRAS) organisms which is safely applied in medical and veterinary functions [1]. Members of LAB share the property of being Gram-positive bacteria that ferment carbohydrates into energy and lactic acid [2]. In addition, LAB produces small organic compounds that give the aroma and flavor to the fermented product [3]. In the food industry, LAB is widely employed as starter cultures and has been indexed as part of human microbiota [4]. Recently, the populace has become more health conscious about the type of diet that they consumed to avoid risk factors that can cause obesity and associated health hazards. Lactobacillus spp. has become increasingly popular and found use as probiotics in foods. “Probiotics are live microorganisms administered in amounts that positively affect the health of the host” [5,6]. Probiotics bacteria are completely non-toxic, have been consumed as part of cultured food such as yoghurt. These beneficial probiotics microorganisms group aid digestion of nutrient substances, they also conserve them from effects of pathogen microorganisms by some substances they revealed while digesting nutrients. LAB is used commonly in milk product which is very important in human development and health [7,8]. Fermented foods are popularly accepted for their flavour, better keeping quality, and the fact that fermentation creates variety among foods.
Yoghurt, cheese and fermented milk products are the main food sources of probiotics [9]. Yoghurt is produced through the fermentation of pasteurized milk by Streptococcus thermophilus and Lactobacillus delbrueckii subsp. Bulgaricus [10]. As the popularity of yoghurt products continues to grow, manufacturers are continuously investigating value-added ingredients to entice health-conscious consumers [11]. The main biochemical changes that occur in cheese manufacture is the production of lactic acid from lactose. This is achieved by different species of lactic acid bacteria (LAB). The responsible flora that form acid development during cheese production are starter cultures that cause decrease in the pH, formation of curd, expulsion of whey [12]. In order to improve the spontaneous traditional fermentation, [13-15] suggested controlled fermentation using mesophilic lactic acid bacteria starter culture. Isolation of lactic acid bacteria (LAB) from fruits and vegetables have frequently been reported [16-18]. Soursop (Annona muricata) fruits are important fruits as they are not only good sources of vitamins, dietary fiber and minerals. They provide flavor, aroma and texture to the pleasures of eating food but also are claimed have anticancer and antioxidant capabilities [19]. Cheese is white soft non-ripened milk made by the addition of a plant extract (Calotropis procera) to the non-pasteurized whole milk from cattle [20]. However, studies on LAB associated with soursop yoghurt in relation to cow and goatmilk yoghurt isolate remain scarce. Therefore, in order to develop a suitable starter for controlled fermentation, isolation and identification of the dominant bacteria involved in the fermentation of milk products and the use of isolates as starter cultures is essential. Moreover, the microorganisms that bring about the natural fermentation of milks from soursop fruit, cowmilk, goatmilk are neither known nor characterize for now. Therefore, the objective of this study was to isolate and characterize lactic acid bacteria from soursop, cowmilk, goatmilk yoghurt and cheese.
Materials and Methods
Sample collection
Yoghurt samples for lactic acid bacteria isolation were prepared from four different samples soursop fruit, cowmilk, goatmilk and cheese respectively.
Pour plate technique: Lactic acid bacteria were isolated from soursop, cowmilk and goatmilk yoghurts and cheese samples using de Man, Rogosa and Sharpe (CM0361 Oxoid, England) according to the method of Dave RI et al. [21]: 10 mL of each sample was taken and homogenized in 90 mL of peptone water. Serial dilutions up to 10-8 were prepared and 1 mL aliquot from 10-5, 10-6, 10-7 dilutions was used for isolation. Each sample was plated twice. The plates were incubated for three days at 37°C under anaerobic conditions (in anaerobe jar using oxoid anaerogen compact). Preliminary identification of lactic acid bacteria was made on the basis of Gram-staining and catalase reaction, followed by microscopic examination to observe cell arrangement and morphological characteristics. After incubation, individual colonies were selected and transferred into sterile broths (MRS or M17 medium for enrichment). The enriched bacteria were purified using streak plate technique. Gram positive and catalase negative cocci and bacilli colonies were stored in glycerol solution (%).
Biochemical identification:
Gram staining procedure: The gram staining procedure was done using crystal violet stain for one minute. The excess stain was removed under tap water. Again, it was stained with gram’s iodine as mordant for one minute and washed under tap water. The washed gram’s iodine mordant was fixed with 5% alcohol for 15seconds and counter-stained with safrainine for 30 sec, washed under tap water and dried with cotton towel gently [21].
Catalase test: Catalase enzymes break down hydrogen peroxide into oxygen and water molecules (2H2O2 → 2H2O + O2) and oxygen production was observed by the generation of O2 bubbles. Catalase test was performed by adding few drops of 3% hydrogen peroxide to a test tube containing 24 hr-old culture of each isolate [21].
Gas production from glucose: The production of carbon dioxide gas from glucose is the test for the determination of homofermentative nature of isolate. MRS broths containing inverted Durham tubes were utilized to test for CO2 production. Five microliters of overnight activated cultures were inoculated into 8 mL MRS containing inverted tubes and incubated for five days at 42°C.
Growth at different temperatures: Growth at 15°C and 45°C are the most frequently used for the classification of bacilli [22]. To determine the growth at given temperatures, the modified MRS media were used. Bromocresol purple was used to determine the colour change in acidity from purple to yellow, indicating lactic acid production and cell growth. Five microliters of overnight activated cultures were inoculated into 5 mL test media, incubated at 45°C and observed for seven days for colour and growth.
Inoculation and isolation of bacteria
MRS (de Man, Rogosa and Sharpe) agar was used for the isolation of lactic acid bacteria. Serial dilutions up to 10-7 were prepared using pour plate technique. Plates were incubated anaerobically in an anaerobic jar (BBL, GasPak) at 32°C for 48 hrs. Bacterial colonies showing clear zones were selected, streaked twice on MRS agar plates for purification and maintained as pure culture over nutrient agar slants (pH 9.0, 4°C). The individual bacterial colonies were stored in 0.8% MRS agar overlaid with glycerol at -20°C. The isolates having clearance zone were selected for colony morphology and some physiological tests. Physiological and biochemical identifications were performed according to the methods and criteria of [23].
Molecular characterization:
DNA isolation: Genomic DNA from all isolates was extracted by using Thottappilly G et al. method [24]. The 16S rRNA gene sequence of LAB was amplified using universal primers EGE1 and L1 from 18-27 bp as forward primer and 1471-1492 bp as reverse primer as described by Pandley KK et al. [25]. The reaction was carried out in Eppendorf Master Cycler. The PCR conditions were standardized as follows: initial denaturation at 94°C for 5 min, 30 cycles of denaturation of 94°C for 30 sec, annealing at 54°C for 30 sec and extension at 72°C for 90 sec and final extension at 72°C for 5 min.
Separation of amplification products and purification of PCR products: 1% of Agarose was dissolved in 100 μL 1x TAE buffer by boiling. It was cooled to 45°C. 15 μL ethidium bromide solution (10 mg/mL), added and stirred. The agarose gel was poured into the gel casting stand and the combs were placed. 3 μL of PCR products were mixed with 3 μL of gel loading dye. The samples were loaded into the wells, starting from the second well on the gel. DNA size- marker (1 kb Fermentas) was loaded into the fist well. PCR products were electrophoreses at 120 V for 1 hr. Amplification products were visualized in a gel documentation system (Vilber-Lormat). The presence of DNA fragments with the size of 1500-2000 bp indicated that the amplification was achieved.
Sequencing the DNA: The sequence data obtained was deposited to NCBI database with BLAST analysis for molecular identification of the organisms.
Results and Discussion
Strains of bacteria isolated from soursop, cowmilk, goatmilk yoghurts and cheese were examined. A total number of 24 isolates were picked from MRS agar plates and were found to belong to the genus Lactobacillus, according to their physiological and biochemical tests. The isolated bacteria were stored in MRS broth culture in temperature -800C for further analysis. The isolates were gram positive when view under light microscope, hence they were gram positive bacteria. They appeared as a chain of 3-4 cells (rounded rods, cocci) in shape, and catalase negative bacteria. All the isolates were subjected to test for gas production from glucose and the test tubes observed for 5 days and there was no gas accumulation in Durham tubes. For identification of bacilli isolates, the growth at 15°C and 45°C were used. However, Lactobacillus delbrueckii ssp bulgaricus cannot grow at 15°C but can grow at 45°C. Their morphology and physiology characteristics are shown in Table 1.
Table 1Biochemical tests for lactic acid bacteria isolates.
| Sample | Gram staining | Cell | Catalase | Growth | Growth at 15°C | Gas from glucose | Glucose | Fructose | Maltose | Sucrose |
|---|---|---|---|---|---|---|---|---|---|---|
| Morphology | at | |||||||||
| 45°C | ||||||||||
| Soursop | G+ | ROD | - | + | - | - | + | + | + | + |
| Yoghurt | ||||||||||
| Cow milk Yoghurt | G+ | ROD | - | + | - | - | + | + | + | + |
| Goat milk Yoghurt | G+ | ROD | - | + | - | - | + | + | + | + |
| Soft Cheese | G+ | ROD | - | + | - | - | + | + | + | + |
Note: G+gram positive; – negative; + positive
Genomic DNA isolation
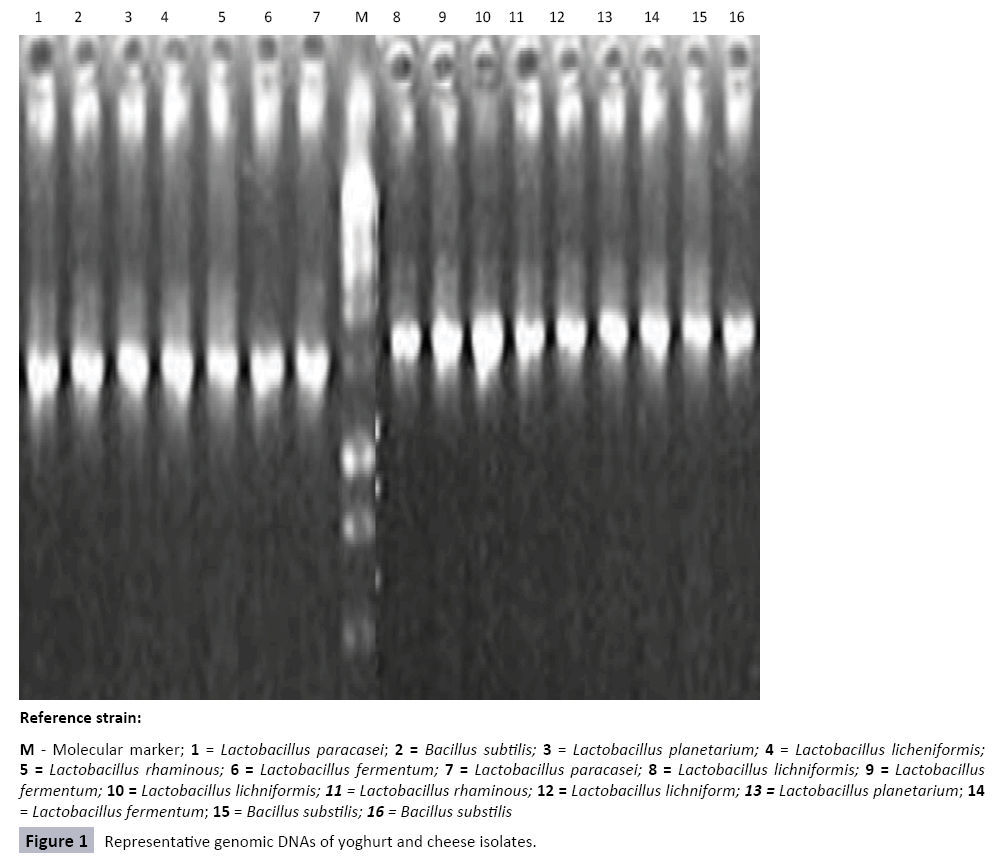
The isolation of genomic DNA was verified with agarose gel electrophoresis. The genomic DNA was seen as a smear of different molecular weight DNAs under UV light. Some genomic DNA patterns of isolates were shown in Figure 1
Molecular characterization of lactic acid bacteria
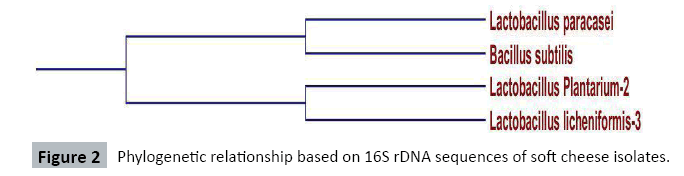
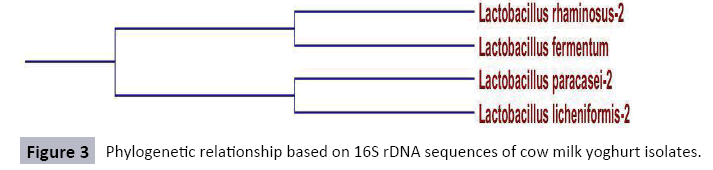
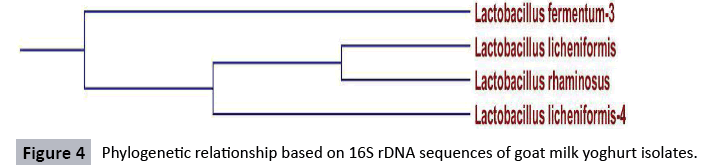
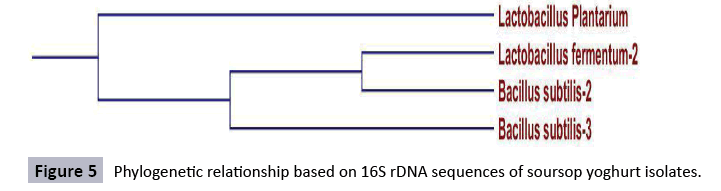
Sequencing results of 16S rDNA was deposited to the database and checked for homology. The sequences blasted on the NCBI gene bank demonstrated 100% homology. The 16S primer was able to amplify DNA isolated from Lactobacillus species. The agarose gel electrophoresis of the PCR products was distinct. The alignment gave us the inter-relationship in the nucleotide arrangement of the organisms. The claude on the phylogenetic tree also demonstrated the relationship of the species organisms (Figures 2-5). The lactic acid bacteria species identified in this study are more diverse as compared to those reported Suleiman AMEH et al. [26] who isolated only two Lactobacillus species (Lactobacillus plantarum and Lactobacillus paracasei) from fermented camel milk (Garris). Such variation in the diversity of lactic acid bacteria observed between cowmilk, goatmilk and soursop yoghurt might be attributed to the inherent characteristics of the fermented milks used and the specific fermentation condition followed. In Rawalpindi region Toqeer A et al. [27] isolated Lactobacillus lactis ssp. cremoris, Lactobacillus lactis ssp. lactis and Lactobacillus acidophilus from camel milk and reported that Lactobacillus acidophilus grew relatively more rapidly in camel milk. Togo MA et al. [28] reported isolation and identification of potential lactic starters from unusual indigenous niche for lactic acid bacteria in opaque beer (Chibuko). Sequences with forward and reverse primers were matched and compared with primer walking and the gaps were completed. The sequence of 16S rDNA analysis of soft cheese isolate is shown in Figure 6. The results of this study revealed that the BLAST analysis at the NCBI gene bank gave 100% homology to Lactobacillus paracasei, 100% homology to Bacillus subtilis, 100% homology to Lactobacillus Plantarium, and 100% homology Lactobacillus lichniformis for cheese isolates. Also, gave 100% homology to Lactobacillus paracasei, 100% homology to Lactobacillus fermentum, 100% homology to Lactobacillus rhaminous, and 100% homology Lactobacillus licheniformis for cowmilk isolates, also gave 100% homology to Lactobacillus fermentum, 100% homology to Lactobacillus licheniformis, 100% homology to Lactobacillus rhaminous, and 100% homology Lactobacillus licheniformis for goatmilk yoghurt and gave 100% homology to Lactobacillus plantarium, 100% homology to Lactobacillus fermentum, 100% homology to Bacillus subtilis, and 100% homology Bacillus subtilis for soursop yoghurt.
Sequencing of soft cheese isolates: The sequences of soft cheese isolates with forward and reverse primers were matched and compared with primer walking and the gaps were completed. The sequences of 16S rDNA product of soft cheese isolate are shown in Figure 6. The BLAST analysis at the NCBI gene bank gaves 100% homology to Lactobacillus paracasei, 100% homology to Bacillus subtilis, 100% homology to Lactobacillus plantarium, and 100% homology to Lactobacillus licheniformis.
The quality of cheese and other fermented food products is dependent on the ability of flavour and aroma production of microorganisms which include starter culture. A number of different LAB has been evaluated for their ability to degrade amino acids to aroma compounds. Lactobacillus lactis subsp. lactis and Lactobacillus lactis subsp. cremoris, Lactobacillius lactis, Lactobacillius helveticus, Lactobacillus bulgaricus, Lactobacillus casei are capable of degrading methionine to methonethiol, dimethyledisulphide and dimethyltrisulphide [29]. The phenotype analysis of the cheese strain indicated its relationship to Lactobacillus plantarum and this strain belongs to the group of the mesophilic lactobacilli; commonly met in the later phase of the maturing of cheese, together with Lactobacillus casei, Lactobacillus brevis and Lactobacillus buchneri [22].
Proteolysis is also an important process that occurs during cheese ripening as lactic acid bacteria uses polypeptides generated by milk clotting enzymes and by bacterial cell-wall proteins. Rennet is the milk clotting enzyme which is responsible for casein degradation and because of casein degradation peptides are produced which are transported into the cell. In the cell, peptidases continue degradation to produce smaller peptides and amino acids. It has been known that amino acid composition plays an essential role in the aroma of cheese [30]. Olarte C, et al. [31] noted that the presence of Lactobacillus plantarum in cheese produced from Cameros goat’s milk decreased the number of the enterobacteria and faecal coliforms in the final product.
C T C G C A A T C T G A T T T T G G C T T C A A T T T T A G T T T
G G G C A A A C C G C A T G A C T T G T A T G T C T T T G A G T
C A C C A A T T G A C G C G T T G A G T T A C T G G A C A C A A
CACCCAGAGCTAACTAATTGCATGATTACCTCAGTTGATGGCACCA
AGGTTGAGAGTGTCGTGAATATGGCGGTCAATATGTATAAAACAA
AAGGGCAACTGCCAGCCACTATTTATGTTGGCTCCGACAATGACCCA
GCTGGTCATCAGCTTTATGACAAGCTGCAAGAATCCATTTTAGCCAA
TACC (Figure 6).
Sequencing of cowmilk yoghurt isolate: Sequencing of cowmilk yoghurt isolate with forward and reverse primers were matched and compared with primer walking and the gaps were completed. On the basis of these key characters, subgroup A1 isolates were identified as Lactobacillus rhaminous-2 and Lactobacillus fermentum while subgroup A2 isolates were identified as Lactobacillus paracasei-2 and Lactobacillus licheniformis-2. The sequences of 16S rDNA product of cow milk yoghurt isolates are shown in Figure 7. The BLAST analysis at the NCBI gene bank gave 100% homology to Lactobacillus paracasei-2, 100% homology to Lactobacillus fermentum, 100% homology to Lactobacillus rhaminous-2, and 100% homology Lactobacillus licheniformis-2. Subgroup A1 and A2 were rod shaped isolates, catalase negative; growth at 45°C, produced carbon dioxide from glucose and both isolates utilised carbohydrates like lactose and maltose. Similar characters for Lactobacillus fermentus have been observed by Charntaraporn P et al. [32]. Lactobacilli are regarded as a major group of probiotic bacteria [33,34]. Salminen S et al. [35] proposed that probiotics are microbial cell preparations or components of microbial cells that have a beneficial effect on the health and well-being of the host. Several lactobacilli, lactococci and bifidobacteria are held to be health-benefiting bacteria [36,37]. The health-promoting effects of LAB are strain specific and result in different mechanisms to produce beneficial health impacts. Lactobacillus rhamnosus GG relieves lactose intolerance symptoms by hydrolyzing lactose into glucose and galactose and forming the physical appearance of milk into a thick substance, such as yoghurt, that passes through the gastrointestinal tract slowly, reducing the lactose pulse in the colon [38-40].
G G G G G A T T T T C A T C G T C G C G C A G G C G T A G T T A
ACTAGCACCGACGGGATGATGGGGACCATGTACCCCACTGTCAACC
CTAGGGCGGGATGGCGGTAGTGGGTTAAGCTGTCCAAAATCTTGTT
TTGCTTAAACTTATGGGAGAGTTTGACGTGCGATAACAGGGCGGCG
ACGGCACAATTGCCCAAAATATTCCCTGCCCAGTTAACCAAAAGAC
CGATCCACGG (Figure 7).
Sequencing of goatmilk yoghurt isolate: Molecular methods are important for bacterial identification [41,42] and possibly more accurate for LAB than are conventional phenotypic methods. In this study, a 1500-2900 bp segment of the 16S rRNA gene of the LAB isolates was sequenced and the sequence compared to strains in the National Center for Biotechnology Information (NCBI) Blast Library (Washington University). Sequencing results revealed that four out of the 10 isolates obtained from the goat milk samples were Lactococcus species. Sequences with forward and reverse primers were matched and compared with primer walking and the gaps were completed. Based on the alignment results, subgroup A1 was found as Lactobacillus fermentus-3, subgroup A2 as Lactobacillus licheniformis-4 while subgroup B1 and B2 as Lactobacillus licheniformis and Lactobacillus rhaminosus. The sequences of 16S rDNA product of goatmilk yoghurt isolates are shown in Figure 8. The BLAST analysis at the NCBI gene bank gave 100% homology to Lactobacillus fermentum-3, 100% homology to Lactobacillus licheniformis, 100% homology to Lactobacillus rhaminous, and 100% homology Lactobacillus licheniformis-4.
Lactic acid bacteria are widely distributed in nature and are representatives of the genera Lactobacillus, Lactococcus, Pediococcus and Leuconostoc. They can be isolated from soils, waters, plants, silages, waste products, from the intestinal tract of animals and humans and they possess stable fermentation characteristics and are resistant to bacteriophages [43]. They have potentials for establishing new, so-called “functional foods” [44,45]. The selection of health-promoting bacteria must rest on the documented impacts of selected probiotic strains on the microbial community harboured within the digestive tract [34]. According to Caplice E et al. [3] LAB produces small organic compounds that give the aroma and flavour to the fermented product: lactic acid bacteria of human and animal origin may serve as bacteria potentially promoting host-specific health and antimicrobial effect exerted by LAB is mainly due to acid production, hydrogen peroxide, fatty acids, aldehydes and other compounds [46]. Hutt P et al. [47] also reported that Lactobacillus strains possess intermediate potency for out-competing cystitiscausing E. coli from the large intestine. Goderska K et al. [48] reported that L. acidophilus gave a growth inhibition zone of 26 mm against H. pylori and E. coli.
A C G T T C A T G G A G T T G G C T A G A C A A A T G T C G G C
GACTTTGGAAAAACTGTGAACGCGCCCTTAACAGCAGTCTTCAAAT
GGTTGTATTGGCCTTCTTGAACATAGTTCTTAAAAGATGCTAGACAG
GTTCTCAACAAACGCCACAGCTTGACCAGTGGCATGATCAATGCCA
AAGTTTCGCGCAGCACCGATGCCACGATGGTCATTAAAACCGGTCG
CGTGAAAAT (Figure 8).
Sequencing of soursop yoghurt isolate: Subgroup A1 isolates of lactic acid bacteria isolated from soursop yoghurt contained Lactobacillus plantarium, subgroup A2 had Bacillus substilis-3 while subgroup B2 contained Lactobacillus fermentus-2 and Bacillus subtilis-2. Lactobacillus planetarium was the only isolate in subgroup A1 category, showing it was abundant in the strain. In the case of tomato fruits, Lactobacillus plantarium was the most abundant species of LAB found [49]. Species such as Weissella cibaria, Lactobacillus plantarium and Lactobacillus lactis subsp. lactis have been frequently found in environments associated with plants [50-52]. Isolations of lactic acid bacteria (LAB) from fruits and vegetables have frequently been reported [18,53]. Bacteriocins produced by LAB have attracted special interest as potential alternative safe commercial food preservatives and LAB have been used as food and feed preservatives for centuries. Bacteriocin-producing LAB can replace chemical preservatives for the prevention of bacterial spoilage and the outgrowth of pathogenic bacteria in food products [51].
The sequences of 16S rDNA product of soursop yoghurt isolate are shown in Figure 9. The BLAST Analysis at the NCBI gene bank gave 100% homology to Lactobacillus plantarium, 100% homology to Lactobacillus fermentum-2, 100% homology to Bacillus subtilis-2, and 100% homology Bacillus subtilis-3. Introduction of Lactobacillus acidophilus in milk makes it more effective in lowering body cholestrol level. In fermented foods, LAB displays numerous antimicrobial activities mainly due to the production of organic acids and other compounds, such as bacteriocins and antifungal peptides [54]. Lactobacillus acidophilus which survives lowest pH ranges and tolerates the bile too has been proved to be very successful in preparation of yoghurt harbouring effective probiotic and these characteristics has made it most effective tool against lactose malabsorption and intolerance [55,56].
References
- Holzapfel WH, Schillinger U (2002) Introduction to pre- and probiotics. J Food Res Int 35: 109-116.
- Jay JM (2000) Fermentation and fermented dairy product. In: Modern food microbiology (6th edn.). Springer, US. pp: 113-114.
- Caplice E, Fitzgerald GF (1999) Food fermentation: role of microorganisms in food production and preservation. Int J Food Microbiol 50: 131-149.
- Sanders ME (2000) Symposium: Probiotic bacteria: Implications for human health: Considerations for use of probiotic bacteria to modulate human health. J Nutr 130: 384S-390S.
- FAO/WHO (2002) Human vitamin and mineral requirements. Report of a Joint FAO and WHO expert consultation, Rome.
- Sanders ME (2003) Probiotics: Considerations for human health. Nutr Rev 61: 91-9.
- Tannock GW, Tisala-Timisjarvi A, Rodtong S, Nq J, Munro K, et al. (1999) Idenitification of Lactobacillus isolates from the gastrointestinal tract, silage and yoghurt by 16S-23S rDNA gene intergenic spacer region sequence comparisons. Appl Environ Microbiol 65: 4264-4267.
- Shinoda T, Kusuda D, Ishida Y, Ikeda N, Kaneko K, et al. (2001) Survival of lactobacillus helticus strain CP53 in the human gastrointestinal tract. Lett Appl Microbiol 32: 108-113.
- Salminen S (1996) Functional dairy foods with Lactobacillus strain GG. Nutr Rev 54: S99-101.
- Tamime AY, Robinson RK (2007) Yoghurt: Science and technology (3rd edn.). Woodhead Publishing Limited, Cambridge. P. 808.
- Allgeyer LC, Miller MJ, Lee SY (2010) Sensory and microbiological quality of yoghurt drinks with prebiotics and probiotics. J Dairy Sci 93: 4471-4479.
- Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM (2002) Recent advances in cheese microbiology. Int Dairy J 11: 259-274.
- Abu-Tarboush HM (1996) Comparison of associative growth and proteolytic activity of yoghurt starters in whole milk from camels and cows. J Dairy Sci 79: 366-377.
- Farah Z, Streiff T, Bachmann MR (1990) Preparation and consumer acceptability tests of fermented camel milk in Kenya. J Dairy Res 57: 281-283.
- Mohamed MA, Larsson-Raznikiewicz M, Mohamud MA (1990) Hard cheese making from camel milk. Milchwissenschaft 45: 716-718.
- Bae S, Fleet GH, Heard GM (2006) Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100: 712-727.
- Chambel L, Chelo IM, Zé-Zé L, Pedro LG, Santos MA, et al. (2006) Leuconostoc pseudoficulneum sp. nov., isolated from a ripe fig. Int J Syst Evol Microbiol 56: 1375-1381.
- Duangjitcharoen Y, Kantachote D, Ongsakul M, Poosaran N, Chaiyasut C (2008) Selection of probiotic lactic acid bacteria isolated from fermented plant beverages. Pak J Biol Sci 11: 652-655.
- Luzia DMM, Jorge N, (2012) Soursop (Annona muricata L.) and sugar apple (Annona squamosa L.): Antioxidant activity, fatty acids profile and determination of tocopherols. Nutr Food Sci 42: 434-441.
- Adeyemi IA, Umar S (1994) Effect of method of manufacture on quality characteristics of kunun-zaki, a millet-based beverage. Nigeria Food Journal 12: 34-41.
- Dave RI, Shah NP (1996) Evaluation of media for selective enumeration of Streptococcus thermophilus, Lactobacillus delbrueckii ssp. bulgaricus, Lactobacillus acidophilus and bifidobacteria. J Dairy Sci 79: 1529-1536.
- Hammes WP, Vogel RF (1995) “The genus Lactobacillus” in the genera of lactic acid bacteria. In B.J.B. Wood. Blackie Academic and Professional, United Kingdom. pp: 19-49.
- Bulut C, Gunes H, Okuklu B, Harsa S, Kilic S, et al. (2003) Homofermentative lactic acid bacteria of a traditional cheese, Comlek peyniri from Cappadocia region. J Dairy Res 71: 19-24.
- Thottappilly G, Thresh JM, Calvert LA, Winter S (2003) Cassava: In virus and virus- like diseases of major crops in developing countries. In: Loebenstein G, Thottappilly G (eds), Kluwer Academic publishers, Netherlands. Pp: 107-165.
- Pandley KK, Mayilaraj S, Chakrabarti T (2002) Pseudomonas indica sp. nov., a novel butane-utilizing spacies. Int J Syst Evol Microbiol 52:1559-1567.
- Suleiman AMEH, Ilayan AA, El-Faki AE (2006) Chemical and microbiological quality of Harris, sudanesse fermented camel’s milk product. Int J Food Sci Technol 41: 321-328.
- Toqeer A, Kanwal R, Athar IH, Ayub N (2002) Isolation and identification of LAB from camel milk. Pak Vet J 22: 141-144.
- Togo MA, Feresu SB, Mutukumiara AN (2002) Identification of lactic acid bacteria isolated from opaque beer (Chibuku) for potential use as starter culture. J Food Technol 7: 93-97.
- Yvon M, Rijnen L (2001) Cheese flavor formation by amino acid catabolism. Int Dairy J 11: 185-201.
- Wouters JTM, Ayad EHE, Hugenholtz J, Smit G (2002) Microbes from raw milk for fermented dairy products. Int Dairy J 12: 91-109.
- Olarte C, Sanz S, Gonzaiez E, Torre P (2000) Identification of lactic acid bacteria isolated from katyk goat. J Appl Microbiol 88: 421- 429.
- Charntaraporn P, Somboon T (2006) Characterisation of lactic acid bacteria from traditional Thai fermented sausages. Journal Culture Collect 5: 46-47.
- Collins JK, Thornton G, Sullivan GO (1998) Selection of probiotic strains for human applications. Int Dairy J 8: 487-490.
- Tannock GW (1998) Studies of the intestinal microbiota: a prerequisite for the development of probiotics. Int Dairy J 8: 527-533.
- Salminen S, Ouwehand A, Benno Y, Lee YK (1999) Probiotics: how should they be defined? Trends Food Sci Technol 10: 107-110.
- Rolfe RD (2000) The role of probiotic cultures in the control of gastrointestinal health. J Nutr 130: 396S-402S.
- Tuohy KM, Probert HM, Smejkal CW, Gibson GR (2003) Review: Using probiotics and probiotics to improve gut health. Drug Discov Today 8: 692-700.
- Hove H, Norgaard H, Brobech MP (1999) Lactic acid bacteria and the human gastrointestinal tract. Eur J Clin Nutr 53: 339-350.
- Heyman M (2000) Effect of lactic acid bacteria on diarrheal diseases. J Am Coll Nutr 19: 137-146.
- Drouault S, Corthier G (2001) Effets des bactéries lactiques ingérées avec des laits fermentés sur la santé. Vet Res 32: 101-117.
- Greetham HL, Giffard C, Hutson RA, Collins MD, Gibson GR (2002) Bacteriology of the Labrador dog gut: A cultural and genotypic approach. J Appl Microbiol 93: 640-646.
- Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, et al. (2002) Molecular diversity of Lactobacillus ssp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 68: 114-123.
- Lee B (1996) Bacteria-based processes and products. In: Fundamentals of Food Biotechnology. VEH, New York. pp: 219-290.
- Reid G (1999) The scientific basis for probiotic strains of Lactobacillus. Appl Environ Microbiol 65: 3763-3766.
- Holzapfel W, Habere P, Geisen R, Bjorkroth J, Schillinger U (2001) Taxonomy and important features probiotic microorganisms in food and nutrition. Am J Clin Nutr 73: 365-373.
- Daeschel MA (1989) Antimicrobial substances from lactic acid bacteria for use as food preservatives. Food Technol 43: 164.
- Hutt P, Shchepetova J, Loivukene K, Kullisaar T, Mikelsaar M (2006) Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero- and uropathogens. J Appl Microbiol 100: 1324-1332.
- Goderska K, Czarnecki Z (2007) Characterisation of selected strains from Lactobacillus acidophilus and Bifidobacterium bifidum. Afr J Microbiol Res 1: 065-078.
- Di-Cagno R, Surico RF, Paradiso A, De Angelis M, Salmon JC, et al. (2009) Effect of autochthonous lactic acid bacteria starters on health-promoting and sensory properties of tomato juices. Int J Food Microbiol 128: 473-483.
- Kostinek M, Specht I, Edward VA, Pinto C, Egounlety M, et al. (2007) Characterisation and biochemical properties of predominant lactic acid bacteria from fermenting cassava for selection as starter cultures. Int J Food Microbiol 114: 342-351.
- Escalante-Minakata P, Blaschek HP, Barba de la Rosa AP, Santos L, De León-Rodríguez A (2008) Identification of yeast and bacteria involved in the mescal fermentation of Agave salmiana. Lett Appl Microbiol 46: 626-630.
- Trias R, Bañeras L, Montesinos E, Badosa E (2008) Lactic acid bacteria from fresh fruit and vegetables as biocontrol agents of phytopathogenic bacteria and fungi. Int Microbiol 11: 231-236.
- Nyanga LK, Nout MJ, Gadaga TH, Theelen B, Boekhout T, et al. (2007) Yeasts and lactic acid bacteria microbiota from masau (Ziziphus mauritiana) fruits and their fermented fruit pulp in Zimbabwe. Int J Food Microbiol 120: 159-166.
- Simova ED, Frengova GI, Beshkova DM (2004) Synthesis of carotenoids by rhodotorula rubra GED8 co-cultured with yoghurt starter cultures in whey ultra filterate. J Ind Microbiol Biotechnol 31: 115-121.
- Mustapha A, Jiang T, Savaiamo DA (1997) Improvement of lactose digestion by humans following ingestion of unfermented acidophilus milk: influence of bile sensitivity, lactose transport and acid tolerance of Lactose acidophilus. J Dairy Sci 8: 1537-1545.
- de Vrese M, Richter B, Fenselau S, Laue C, Schrezenmeir J, et al. (2001) Probiotics-compensation lactase insufficiency. Am J Clin Nutr 73: 421S-429S.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences