Comparative Expression Analysis of Ghrelin Hormone in Five Meat-Genotype Broiler Chickens
1Department of Animal Production, Faculty of Agriculture, Cairo University, Giza, Egypt
2Department of Animal and Avian Sciences, University of Maryland, College Park, USA
- *Corresponding Author:
- Mona M Ghaly
Department of Animal Production, Faculty of Agriculture,
Cairo University, Gamma St.,
Giza, Egypt
Tel: 00201002828243
E-mail: Monaghaly19@yahoo.com
Received date: August 03, 2018; Accepted date: August 09, 2018; Published date: August 17, 2018
Citation: Ghaly MM, Li M (2018) Comparative Expression Analysis of Ghrelin Hormone in Five Meat-Genotype Broiler Chickens. J Genom Gene Study Vol.1 No.1:4
Abstract
The present investigation aims to study the Ghrelin gene (GHRL) expression patterns of meat-type hybrids, Aviagen, Arbor Acres, Hubbard, Cobb and Ross at 21 and 37 days of age. There was no association between the level of expression and either phenotypic or carcass parameters for all genotypes at 21 days. The highest transcript abundance of Gherlin mRNA for Ross genotype had a significant negative association with weight gain (WG 3-5 wks), growth efficiency (GE 0-5 and 3-5 wks), specific growth rate (SGR 0-37 and 21-37 day), carcass weight (CW), shank length (SL) and wing weight (WW). Aviagen hybrid, the lowest gene expression of all, had high significant positive association for both abdominal fat and abdominal fat % at slaughter age. Arbor Acre hybrid had significant (p ≤ 0.05) negative association with fat % of breast muscle meat by. The drumstick muscle fat % of Hubbard hybrid chickens was negatively regressed on the Gherlin mRNA expression. These findings suggested that the differences in transcript level and response among resembles of meat type hybrids might be the result of a different genetic origin for them and further investigation is recommended for Aviagen and Ross hybrids.
Keywords
Gene expression; Ghrelin gene; Growth performance; Meat type chickens
Abbreviations
GHRL: Ghrelin Gene; GHSR: Secretagogue Receptor; GE: Growth efficiency; RC: Regression coefficient; SGR: Specific Growth Rate
Introduction
Ghrelin first discovery in mammalian species and identified in rat stomach [1]. The proventriculus Ghrelin peptide was isolated for the first time from chicken and 26 amino acids were found compared to 28 in mammals with 54% of similarities to mammal’s Ghrelin [2]. Ghrelin has recently been distinguished in birds like Chickens, Ducks, Japanese quail, Emu, Goose, Turkey [3]. Ghrelin is predominantly a product of gastric tissue but its expression has been detected in many tissues including brain, hypothalamus, intestine, cells of the immune system, thyroid gland, kidney, lung, pancreas, pituitary and placenta [4,5]. Chicken Ghrelin gene is primarily present in the proventriculus but absent in the gizzard [6]. Literatures have showed significant functions for Ghrelin as a member of G-protein-coupled receptor family.
Main function difference in Ghrelin between birds and mammals reflects its role in feed intake regulation. Several reports showed that the injections of acylated Ghrelin peptide increase plasma corticosterone levels [2,7,8]. The Ghrelin neural network in the paraventricular nucleus (PVN) is complex because of Ghrelin neurons send efferent fibers onto neuropeptide Y (NPY) neurons. As a result, gamma-Amino butyric acid (GABA) release is suppressed, corticotrophin-releasing hormone (CRH)-expressing neurons is stimulated and finally, adrenocorticotrophic hormone (ACTH) and cortisol are released [9]. The acylated Ghrelin peptide injections elevate plasma corticosterone levels [2,7] and suppress feeding behavior in chickens [2,7,10-12]. This finding confirms earlier evidence suggesting that Ghrelin is an anorixegenic hormone, decreases appetite in avian species in chickens [8,10,11,13,14]. Ghrelin was reviewed as an orexigenic in all birds [15,16]. One of the pathways for the anorexigenic effect of high ambient temperature in laying hens might be mediated by its effects on the hypothalamic and gastrointestinal Ghrelin signals [17]. However, Ghrelin may be anti-lipogenic in birds [11,18] and it significantly increases metabolic rate, lipolysis and weight loss in chickens [19]. Acylated Ghrelin concentrations of garden warblers (Sylvia borin) with larger fat stores were higher than those of birds without fat stores, Further, injections of unacylated Ghrelin decreased food intake and increased migratory restlessness of wild birds [20]. Different dietary energy and protein levels have no effect on Ghrelin gene mRNA expression for broiler chicken [21]. Passive immunization against Ghrelin in turkeys was associated with a significant increase in feed consumption [22]. It is suggested that the loss of anorexigenic Ghrelin is a predatory adaptation that results in increased food-seeking behavior and feeding in falcons [23]. In mammals however, Ghrelin is associated with a positive energy balance and lipogenesis. In addition, high doses of Ghrelin in humans increase ACTH, prolactin, and cortisol levels [24], which may explain the amenorrhea and behavioural changes observed in Anorexia nervosa (AN) patients. The purpose of this study is to figure out and explore the Ghrelin gene expression as a candidate gene responsible for both growth performance and fatness, and compare how its association with both growth traits and fatness among five meat-type hybrids.
Materials and Methods
Experimental birds, diets and tissue sampling
This experiment was conducted at the Poultry Research Station, Faculty of Agriculture, Cairo University, Egypt. In this study, five broiler genotypes including Aviagen, Arbor Acre, Hubbard, Cobb and Ross were used. A total of 5000 broiler chicks (1000 per genotype) were tested. They were fed ad-libitum on the standard starter diets for 14 days and then received standard growing diet up to 37 days of age. Birds were allocated in equal numbers in individual cages and daily exposed to 16 h light and 8h dark cycle under the same environmental conditions with access to feed and fresh tap water all over the day during the experimental period. The body weights (BW) were repeatedly recorded until the age of 37 days of age. Total of 50 birds (6 individuals × 5 genotypes) were slaughtered by cervical dislocation at 37 days of age and (4 individuals × 5 genotypes) for 21 days of age. Proventriculus was aseptically removed and placed in RNA later and kept at -80°C until analysis. Carcass traits were measured for the same slaughtered chickens at 37 days of age, which including carcass weight (CW/g), fore half (FH), major and small pectoral muscle weight (MMP/g) and (SMP/g), thigh muscle weight (TMW/g), drumstick muscle weight (DMW/g), shank length (SL/cm), head weight (HW/g), neck weight (NW/g), edible parts weights/g (heart, liver, spleen and gizzard), abdominal fat (AF/g) and wing weight (WW/g). Relative measures according to the carcass weight were also recorded. Growth efficiency and specific growth rate were calculated [25]. Feed conversion ratio (FCR) was obtained by dividing the total feed intake of each bird by the total grams of live body weight of bird at slaughter age.
Gene expression analyses
RNA extraction and reverse transcription-PCR assay for Ghrelin gene expression: Total of 0.5 g from chicken proventriculus tissues was removed after slaughter and placed in RNA later solution and kept at -80°C until the time of analysis. Total mRNA was isolated by using Qiazol (Rneasy tissue mini kit) reagent procedure, Qiagen according to the manufacturer’s instructions (Qiagen, Germany). The quantity and integrity of isolated RNA were determined for each sample by using using NanoDropTM 2000 Spectrophotometer-Thermo Scientific Inc (Wilmington, Delware- USA). Then RNA samples were stored at -80°C until use. Reverse Transcription Polymerase Chain Reaction (RT-PCR) was performed by using a HIGH CAPACITY cDNA reverse transcription kit containing RNA (1 μg) and 20 pmol gene-specific primer, 9700 GeneAmp PCR-Applied Biosystems (California, USA). The mixture was incubated at 25°C for 10 minutes for enzyme activation, 37°C for 120 minutes, 85°C for deactivation for the enzyme, and then stored at -20°C. A chicken Ghrelin fragment (203 bp) was amplified with a sense primer ( 5- ׳ CCT TGG GAC AGA AAC TGC TC- ׳ 3 ) and an anti-sense primer ( 5- ׳ CAC CAA TTT CAA AAG GAA CG- 16 ] (׳ 3 ]. Chicken ribosomal 18S RNA was chosen as a reference gene. (Fragment size: 148 bp): Sense primer ( 5- ׳ CGC GTG CAT TTA TCA GAC CA-3 ׳) and an anti-sense primer ( 5- ׳ ACC CGT GGT CAC CAT GGT A-3 ׳), (Primer- Invetrogen, USA).
Real-time PCR testing on mRNA level in proventriculus: Ghrelin mRNA quantitation in proventriculus tissue by real-time RT-PCR using a master mix containing SYBRTM Green PCR Master Mix- Life Technologies (California, US). Ten pmol forward primer, 10 pmol reverse primer, cDNA, water was prepared to perform realtime PCR. The following PCR protocol was used on the 500 Real- Time PCR System-Applied Biosystems TM (California USA). Initial steps contain 2 minutes at 50°C and 10 minutes at 95°C, followed by two-step amplification program (15 seconds at 95°C followed by 1 minute at 61°C) repeated 45 times. Runs were performed in three technical replicates per sample.
Statistical analysis: Expression levels of mRNA were measured as cycle threshold values for each gene relative to its cycle threshold values for ribosomal 18S RNA (housekeeping gene). The relative quantification was calculated with the following equations: ΔCt=CtGhrelin-Ct18S; After all the ΔCt values were obtained for all biological and technical replicates, the mean ΔCt values for each genotype five weeks was compared to the mean ΔCt for the three weeks as a calibrator using the 2-ΔΔCT method. Thus, all the five genotypes data five weeks for Ghrelin is expressed as the fold-difference relative to calibrator three weeks. The amount of target molecules relative to the calibrator was calculated by 2-ΔΔCT method. Data was analyzed using SAS (2004). The model included genotype and age as main fixed effects; the individual bird was the experimental unit for gene expression analysis. Gene expression-trait association analysis was performed by SAS GLM procedure.
Gene expression-phenotype association analysis and contrast were performed by SAS GLM procedure. The genetic effects were analyzed by fixed procedure according to the following model: Y=μ+G+A+e, where Y=an observation on the trait, μ=the overall population mean, G= the fixed effect of genotype, A=the fixed effect of age and e=the residual random error. The significant associations were calculated using simple linear regression as the following model: Y=b0+b1X+e where Y=the dependent phenotypic variable, X=the independent target gene expression variable deviated from its housekeeping gene, b0=the intercept and b1=the association of gene effect and e=the residual random error. Clustering procedures used to calculate nearest neighbour hierarchical method by computer program SAS 9.1.
Results and Discussion
Expression of Ghrelin mRNA level in chicken proventriculus by real-time quantitative PCR
Least squares analysis of variance
Three weeks of age: RT-PCR was performed to detect the Ghrelin gene expression patterns between individuals from five meat chickens genotypes, Aviagen, Arbor Acre, Hubbard, Cobb and Ross. As presented in Table 1, expression levels among genotypes showed that Ross genotype recorded the lowest ΔCT mean (12.22) (highest expression) of all. Least squares means of ΔCT for other genotypes, Aviagen, Arbore Acres, Hubbard and Cobb are significantly the same.
Table 1: Least squares mean of Gherlin ∆CT ± standard errors for different genotypes pairs and genotype contrasts versus the rest (linear function ± SE) at 3-weeks of age.
n=Number of missed replicate; ∆CT1=It represent significance within column with different super alphabetic.
| Versus the rest | p-Value | F-Value | Contrast SS | Estimate | Versus | ∆CT1 | Genotype {4(3rep-n)} | |||
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | F-Value | Contrast SS | Estimate | |||||||
| 0.9057 | 0.01 | 0.2365568 | 0.170 ± 1.43 | 0.811 | 0.06 | 0.9636581 | -0.428 ± 1.784 | Arbor Acre | 17.03 ± 1.291 | Aviagen (11) |
| 0.2229 | 1.52 | 25.4190466 | -2.254 ± 1.826 | Hubb | ||||||
| 0.5713 | 0.32 | 5.4181214 | -1.017 ± 1.78 | Cobb | ||||||
| 0.0177 | 6.04 | 100.6648373 | 4.383 ± 1.784 | Ross | ||||||
| 0.6117 | 0.26 | 4.3542556 | 0.706 ± 1.38 | 0.811 | 0.06 | 0.9636581 | -0.428 ± 1.784 | Aviagen | 16.60 ± 1.291 | Arbor Acre (10) |
| 0.3113 | 1.05 | 17.4616573 | -1.825 ± 1.784 | Hubb | ||||||
| 0.737 | 0.11 | 1.9023723 | -0.588 ± 1.741 | Cobb | ||||||
| 0.0081 | 7.64 | 127.3932134 | 4.812 ± 1.741 | Ross | ||||||
| 0.0424 | 4.35 | 72.470515 | 2.98 ± 1.43 | 0.2229 | 1.52 | 25.4190466 | -2.254 ± 1.826 | Aviagen | 17.62 ± 1.231 | Hubb (11) |
| 0.3113 | 1.05 | 17.4616573 | 1.825 ± 1.784 | Arbor Acre | ||||||
| 0.4912 | 0.48 | 8.0241318 | 1.237 ± 1.784 | Cobb | ||||||
| 0.0005 | 13.85 | 230.8443412 | 6.638 ± 1.784 | Ross | ||||||
| 0.3024 | 1.09 | 18.1192568 | 1.442 ± 1.38 | 0.5713 | 0.32 | 5.4181214 | -1.017 ± 1.784 | Aviagen | 18.86 ± 1.291 | Cobb (11) |
| 0.737 | 0.11 | 1.9023723 | 0.588 ± 1.741 | Arbor Acre | ||||||
| 0.4912 | 0.48 | 8.0241318 | -1.237 ± 1.784 | Hubb | ||||||
| 0.0032 | 9.62 | 160.4307311 | 5.400 ± 1.741 | Ross | ||||||
| 0.0004 | 14.73 | 245.5752531 | -5.308 ± 1.38 | 0.0177 | 6.04 | 100.6648373 | 4.383 ± 1.784 | Aviagen | 12.22 ± 1.231 | Ross (11) |
| 0.0081 | 7.64 | 127.3932134 | -4.812 ± 1.741 | Arbor Acre | ||||||
| 0.0005 | 13.85 | 230.8443412 | -6.638 ± 1.784 | Hubb | ||||||
| 0.0032 | 9.62 | 160.4307311 | -5.400 ± 1.74 | Cobb | ||||||
Five weeks of age: RT-PCR for Gherlin mRNA assay showed that as presented in Table 2 both Aviagen and Arbore Acres showed significantly the highest least squares means of ΔCT expression among genotypes (lowest expression), (17.71 and 17.27, respectively). Ross genotype recorded the lowest ΔCT mean (14.87), (highest expression) of all. Meanwhile, both Hubbard and Cobb recorded intermediate significant different scores of all.
Table 2: Least squares mean of Gherlin ∆CT ± standard errors for different genotypes pairs and genotype contrasts versus the rest (linear function ± SE) at 5-weeks of age.
n=Number of missed replicate; ∆CT1=It represent significance within column with different super alphabetic.
| Versus the rest | p-Value | F-Value | Contrast SS | Estimate | Versus | ∆CT1 | Genotype {6 (3rep-n) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | F-Value | Contrast SS | Estimate | |||||||
| 0.0423 | 4.25 | 15.5768514 | 1.043 ± 0.507 | 0.5051 | 0.45 | 1.64019271 | -0.440 ± 0.657 | Arbor Acre | 17.71 ± 0.478 | Aviagen (16) |
| 0.0104 | 6.88 | 25.17246141 | 1.672 ± 0.637 | Hubb | ||||||
| 0.4053 | 0.7 | 2.56143161 | 0.533 ± 0.637 | Cobb | ||||||
| 0.0003 | 14.25 | 52.15900068 | 2.407 ± 0.637 | Ross | ||||||
| 0.0034 | 9.08 | 33.2348859 | 1.593 ± 0.507 | 0.5051 | 0.45 | 1.64019271 | -0.440 ± 0.657 | Aviagen | 17.27 ± 0.450 | Arbor Acre (18) |
| 0.0019 | 10.32 | 37.79931862 | 2.112 ± 0.657 | Hubb | ||||||
| 0.1424 | 2.19 | 8.0279405068.67 | 0.973 ± 0.657 | Cobb | ||||||
| <.0001 | 18.76 | 742734 | 2.847 ± 0.657 | Ross | ||||||
| 0.0415 | 4.29 | 15.6933955 | -1.047 ± 0.507 | 0.0104 | 6.88 | 37.79931862 | 1.672 ± 0.637 | Aviagen | 16.74 ± 0.450 | Hubb (18) |
| 0.0019 | 10.32 | 25.17246141 | 2.112 ± 0.657 | Arbor Acre | ||||||
| 0.0778 | 3.19 | 11.674311654.86 | -1.138 ± 0.637 | Cobb | ||||||
| 0.2525 | 1.33 | 159437 | 0.734 ± 0.637 | Ross | ||||||
| 0.4588 | 0.55 | 2.0280427 | 0.376 ± 0.507 | 0.4053 | 0.7 | 2.56143161 | 0.533 ± 0.637 | Aviagen | 15.60 ± 0.450 | Cobb (18) |
| 0.1424 | 2.19 | 8.0279405 | -0.973 ± 0.657 | Arbor Acre | ||||||
| 0.0778 | 3.19 | 11.67431165 | 1.138 ± 0.637 | Hubb | ||||||
| 0.0043 | 8.63 | 31.60319083 | 0.734 ± 0.637 | Ross | ||||||
| 0.0002 | 15.11 | 55.3075118 | -1.965 ± 0.507 | 0.0003 | 14.25 | 52.15900068 | 2.407 ± 0.637 | Aviagen | 14.87 ± 450 | Ross (18) |
| <.0001 | 18.76 | 68.67742734 | -2.847 ± 657 | Arbor Acre | ||||||
| 0.2525 | 1.33 | 4.86159437 | -0.734 ± 0.637 | Hubb | ||||||
| 0.0043 | 8.63 | 31.60319083 | -1.873 ± 0.637 | Cobb | ||||||
Linear contrasts in two-way analysis of variance
At 3-weeks of age: non-significance pair’s differences between Aviagen, Arbor Acres, Hubbard and Cobb were shown as revealed by linear contrasts coefficients (Table 1). Only Ross genotype had significant pair linear coefficient difference with others. Over the rest contrast, only Hubbard genotype recorded highest ΔCT least squares means over the rest (+ 2.98) and become significantly superior over the rest (p 0.0424). Ross genotype recorded lowest ΔCT least squares means than the rest (-5.308) and become significantly lower than the rest (p 0.0004).
At 5-weeks of age: Ross, the lowest significant ΔCT mean of all genotypes seemed to be not significant from Hubbard genotype (p=0.2525) although they are significantly separated by analysis of variance. Also contrast showed a non- significance difference between both Arbor Acres and Cobb (p=0.1424), although least square means by analysis of variance observe a significant difference as shown in Table 2. The same relationship was observed between both Aviagen and Cobb. Contrast analysis confirms the non-significance difference between Aviagen and Arbor Acres. Contrast over the rest revealed that Aviagen and Arbore Acres genotypes recorded highest ΔCT least squares means over the rest (+1.043 and +1.593), respectively and become significantly superior over the rest (p 0.0423 and 0.0034), respectively. Hubbard and Ross genotype recorded lowest ΔCT least squares means against the rest (-1.047 and -1.965), respectively, and become significantly retarded than the rest (p 0.0415 and 0.0002), respectively. Cobb genotype, the unique that represent non- significant ΔCT mean (0.376) over the rest. As reported by the authors [26,27] contrasts observe comparisons among groups and show difference between specific pairs of groups obviously.
Genotypes allocation to clusters and cluster distance
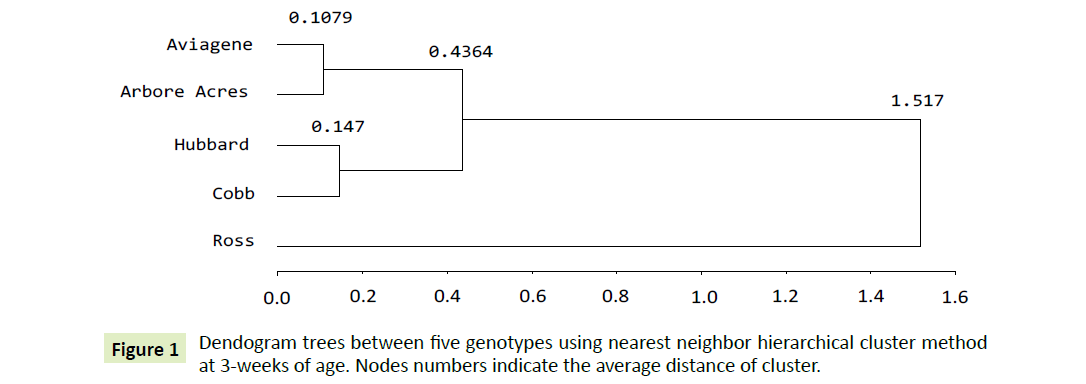
Three weeks: Phylogenetic tree for five independent genotype populations by the nearest neighbour method shows three distinct clusters. The first one aggregates the, Aviagen and Arbore Acres genotypes as in Figure 1. Their least square mean are significantly the same (Table 1), they seems to be the most homologous groups. Their inter cluster distance was lowest among all clusters (0.1079). The second cluster compresses Hubbard and Cobb genotypes at 0.147 point of distance. The third one, aggregate the first and the second clusters in a nearest third nest at 0.4364 degree of distance. Finally, Ross is far away from the four genotype by 1.0806 point. Dissimilarity between genotypes reflects the existence of low gene flow between them [28].
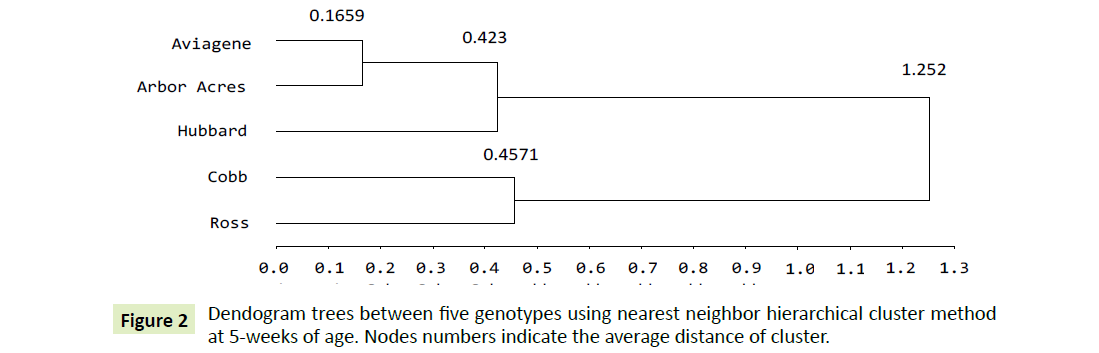
Five weeks: The dendogram clustering procedures at five weeks of age splits the genetic divergence between the five genotypes in three distinct clusters as in Figure 2. The first cluster grouped Aviagen and Arbor Acre as the nearest homologous group confirming having a significant similar pattern for non-significant ΔCT least squares means as revealed in Table 2. Also they both recorded a highest significant ΔCT least squares means over the rest genotypes (+1.043) and (+1.593), respectively. Their inter cluster distance was lowest among all clusters (0.1659). The second one, Hubbard, is much close to the first nest and had joined to it at (0.423) points. Cobb and Ross genotypes had merged in the third cluster. Their ΔCT least squares means are significantly the same Table 2. The intra cluster distance was found to be (0.4571) reflects a high variation exists among them [28]. Finally, the three clusters, aggregates the five genotypes at 1.252 point confirming the existence variation between them.
Fold change profile of Ghrelin mRNA level in proventriculus for five weeks over three weeks
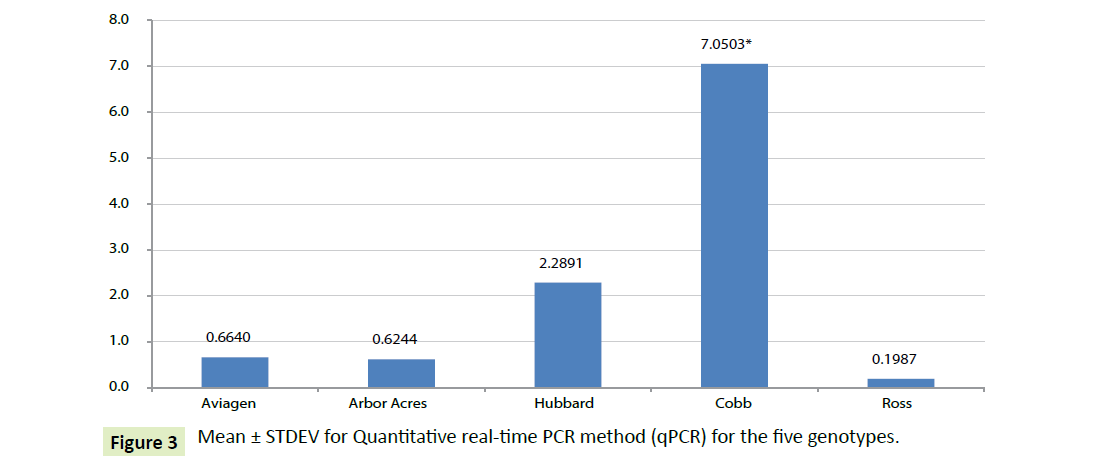
For each genotype, the gene expression profile of three weeks of age was used as control (calibrator) for 5 weeks of age and fold change analyses is shown in Figure 3. Depending on Table 3, the expression of the Cobb Ghrelin mRNA 5-weeks of age had significantly elevated 7 folds than that of three weeks where ΔCT was significantly reduced by age (-3.254). Hubbard genotype Ghrelin mRNA elevated by 2.289 at older age, but not significant by age. Other genotypes their Ghrelin mRNA expression scored non-significant retardation by age, the lowest retardation was for Ross genotype (0.1986).
Table 3: Five weeks contrast versus three weeks for Gherlin mRNA ∆CT (linear function ± SE) for five genotypes and its Fold change expression calculated by ΔΔCT method.
| Genotype | Estimate | ∆∆CT | Average ∆∆CT | Fold Change |
|---|---|---|---|---|
| ∆CT 5 wks-∆CT 3 wks | ||||
| Aviagen | 0.683 ± 1.134NS | 0.591 ± 1.874 | 0.591 (-1.284 to 2.466) | 0.6639507 |
| Arbor Acres | 0.672 ± 0.333NS | 0.679 ± 1.146 | 0.679 (-0.467 to 1.825) | 0.62444169 |
| Hubbard | -0.878 ± 1.011NS | -1.194 ± 2.282 | -1.194 (-3.476 to 1.087) | 2.28908328 |
| Cobb | -3.254 ± 1.328 | -2.818 ± 10.638 | -2.818(-13.46 to 7.820) | 7.05031524 |
| Ross | 1.619 ± 1.283 NS | 2.332 ± 1.519 | 2.332(-0.187 to 4.850) | 0.19866653 |
Association between the mRNA expression and phenotypic traits at 21 days of age
There was no association between the level of Ghrelin mRNA expression and all studied phenotypic growth parameters at 21 days of age for all hybrid genotypes.
Association between the mRNA expression and both growth and carcass traits at slaughter
As listed in Tables 4 and 5, there were no significant association between GHRL mRNA expression levels and most of the studied growth and carcass traits for Aviagen, Arbor Acre, Hubbard and Cobb hybrids except the Ross genotype, the significantly had the lowest ΔCT mean of all (highest expression). The endogenous Ghrelin seems to inhibit chicken growth for most of its recorded growth parameter traits (Table 4). The Ross hybrid, the highest gene expression, observed a highly significant (p ≤ 0.01) negative association between the level of expression and weight gain 21-37 day by -83.952, significant negative (p ≤ 0.05) slop for growth efficiency 0-37 days (-2.406), growth efficiency 21-37 days (-0.457), specific growth rate 0 - 37 days (-0.011), specific growth rate 21-37 days (-0.026) as in (Table 3). Also as presented in Table 4, carcass weight (-603.009), shank length (-1.402) and wing weight (-58.748). This is suggested that Ghrelin plays negative effect on chicken growth for that hybrid. Only for the Arbor Acre hybrid, the Specific Growth rate 0-37 (-0.005) (Table 3), and the drumstick weight traits had a negative significant regression on the Ghrelin expression as expressed by the negative slop of (-16.92) (Table 5). On the contrary, it seems that the lower expression in Aviagen hybrid as in (Table 4), had significant (p ≤ 0.05) positive association with the weights at early ages 14 days and 28 days by about 7.727 and 123.856, respectively. Genotype with the lowest Ghrelin mRNA was the predominant for chicken growth [29]. Growth performance and carcass traits varied among the different five meat type hybrids according to their different Ghrelin mRNA expression. Similar studies showed significant differences between genotypes of Ghrelin gene on some growth traits of TC and RW chicken breeds [30-32]. The growth and body composition traits are important for livestock production. However, these traits are controlled by poly-genes and according to the major gene model; a few genes may be responsible for a relatively large proportion of the genetic variation. Furthermore, these genes may be linked to some quantitative trait loci that may be associated with growth and body composition traits of animals. Most of the variations in the gene are single nucleotide polymorphisms (SNPs). A single SNP can greatly affect the performance traits. For example, the sex-linked dwarf allele in chickens is a single nucleotide mutation at an exon-intron junction of the GH receptor gene GHR [33]. A QTL for muscle growth in pigs was caused by a nucleotide substitution in intron 3 of the insulin-like growth factor 2 gene [34]. In chickens, a total of 37 SNPs and one 6 bp indel were detected in the full length of the GHSR gene. Some of these SNPs were associated with chicken fatness and muscle fiber traits [35]. Similarly, the presence or absence of specific allele’s formation leads to potential correlations between the gene polymorphisms and economic traits in chickens [29]. In chickens and ducks, the several polymorphisms were found in GHRL gene and some of them were scientifically associated with abdominal fat weight, crude protein content of leg muscle and subcutaneous fat thickness [36]. In pigs, the mutation located in the promoter region affected expression of the GHRL gene that leads to a significant (p ≤ 0.05) different expression level between three genotypes [37]. Perhaps, the existences of SNPs influence on splicing and express of Ghrelin gene and its function in chickens.
Table 4 Regression coefficient (RC ± SE) differences for gene expression on growth traits between chickens with different genotypes at 37-days of age.
BW: Body Weight; WG: Weight Gain; GE: Growth Efficiency; SGR: Specific Growth Rate
| Genotype | Avian | Arbor Acre | Hubbard | Cobb | Ross |
|---|---|---|---|---|---|
| BW 14-d | 7.72 ± 4.84 | -13.60 ± 35.35 | -4.940 ± 13.73 | 13.13 ± 54.85 | -2.44 ± 90.32 |
| BW 28-d | 123.85 ± 38.40 | -23.51 ± 75.56 | -29.22 ± 32.37 | -61.87 ± 19.08 | -29.94 ± 193.28 |
| WG 21-37d | 48.32 ± 42.17 | -15.68 ± 100.95 | 21.42 ± 20.48 | 0.81 ± 119.09 | -83.95 ± 60.68 |
| GE 0-37d | 0.40 ± 0.56 | -1.07 ± 0.50 | 0.245 ± 0.29 | -1.162 ± 1.03 | -2.41 ± 0.56 |
| GE 21-37d | 0.062 ± 0.04 | -0.13 ± 0.16 | 0.07 ± 0.06 | 0.11 ± 0.13 | -0.46 ± 0.13 |
| SGR 0-37d | 0.002 ± 0.002 | -0.005 ± 0.001 | 0.002 ± 0.001 | -0.007 ± 0.004 | -0.011 ± 0.00 |
| SGR 21-37d | 0.004 ± 0.002 | -0.009 ± 0.01 | 0.005 ± 0.003 | 0.000 ± 0.005 | -0.026 ± 0.01 |
| FCR | 0.018 ± 0.004 | 0.013 ± 0.011NS | -0.004 ± 0.003NS | -0.006 ± 0.012NS | -0.067 ± 0.022 |
Table 5 Regression coefficient (RC ± SE) differences for gene expression on carcass traits between chickens with different genotypes at 37-days of age.
CW: Carcass Weight; LW: Leg Weight; SHL: Shank Length; WW: Wing Weight; DW: Drumstick Weight (g); FH: Fore Half; SP: Small Pectorals; AF: Abdominal Fat; BF: Brest Fat, DF: Drumstick Fat; RC: Regression Coefficient; SE: Standard Error.
| Traits | Genotype | ||||
|---|---|---|---|---|---|
| Avian | Arbor Acre | Hubbard | Cobb | Ross | |
| CW/g | 83.57 ± 84.29 | -05.14 ± 164.88 | 47.30 ± 38.34 | -114.57 ± 205.80 | -603.01 ± 215.6 |
| LW/g | -3.41 ± 8.14 | -18.97 ± 15.85 | 8.71 ± 3.17 | -1.04 ± 19.48 | -37.07 ± 15.32 |
| SL/cm | -0.07 ± 0.33 | -0.678 ± 0.65 | 0.298 ± 0.21 | 0.213 ± 0.71 | -1.40 ± 0.43 |
| WW/g | 1.640 ± 7.42 | -13.03 ± 23.66 | 4.54 ± 3.36 | -5.06 ± 28.72 | -58.75 ± 14.95 |
| DW/g | -0.73 ± 4.70 | -16.92 ± 5.84 | 1.21 ± 2.93 | -12.88 ± 10.96 | -22.48 ± 14.46 |
| DW % | -0.004 ± 0.004 | -0.01 ± 0.002 | -0.001 ± 0.002 | -0.01 ± 0.003 | 0.004 ± 0.01 |
| FH/g | 0.049 ± 0.01 | -82.57 ± 63.31 | -0.002 ± 0.004 | -0.048 ± 0.04 | -0.023 ± 0.19 |
| SP% | 0.001 ± 0.001 | 0.001 ± 0.00 | -0.001 ± 0.00 | -0.002 ± 0.0005 | 0.001 ± 0.002 |
| Heart/g | -0.64 ± 0.54 | -2.48 ± 1.20 | 0.781 ± 0.17 | 0.121 ± 0.07 | -4.73 ± 1.71 |
| Head% | -0.01 ± 0.001 | 0.000 ± 0.003 | 0.000 ± 0.002 | -0.007 ± 0.02 | -0.008 ± 0.01 |
| Neck% | 0.005 ± 0.005 | 0.005 ± 0.006 | 0.000 ± 0.003 | 0.006 ± 0.02 | 0.021 ± 0.005 |
| AF/g | 15.01 ± 2.31 | -2.92 ± 3.24 | -6.99 ± 3.22 | -25.41 ± 15.26 | -22.15 ± 9.86 |
| AF % | 0.012 ± 0.001 | 0.003 ± 0.01 | -0.01 ± 0.003 | -0.021 ± 0.01 | -0.002 ± 0.01 |
| BF % | 0.05 ± 0.09 | -0.39 ± 0.11 | 0.008 ± 0.07 | 0.087 ± 0.36 | -0.044 ± 0.25 |
| DF % | 0.83 ± 0.62 | -2.65 ± 2.34 | -2.01 ± 0.31 | 1.01 ± 1.45 | 0.87 ± 0.74 |
Association between the Ghrelin mRNA expression and fatness traits
Interestingly, for fatness, in Aviagen hybrid, the highest ΔCT mean (lowest gene expression) of all (Table 2), had high significant positive association for both abdominal fat and abdominal fat % by about 15.0117 and 0.0118, respectively (Table 5) at slaughter age. Meanwhile, Arbor Acre, the lowest ΔCT mean (the highest) expression of the gene in Table 2 had significant negative association with fat % of breast muscle meat by (-0.395) (Table 5). The drumstick muscle fat % of Hubbard hybrid chickens was negatively regressed on the ΔCT mean GHRL mRNA expression by (-2.013) as in Table 5. These findings are consistent with other researches whom concluded that Ghrelin hormone may be antilipogenic in birds and with the Ghrelin inhibits fat build-up in birds by decreasing lipogenic enzyme fatty acid synthase (FAS) in the liver, the major site for building up fat stores in birds and significant increase in metabolic rate [11,18,19].
Association between the Ghrelin mRNA expression and feed conversion ratio
There is significant difference among the five hybrids for their feed conversion ratios (FCR) at 37 days of age (Table 5). Although there is neither significant difference among them for live body weight nor total feed consumption per bird up to the same age (Ghaly, M. Unpublished data). It is noticed that the reduction of Ghrelin mRNA expression for Aviagen genotype (highest ΔCT mean) was accompanied by a significant positive regression on FCR ratio (0.018). It is supposed that the shortage presence of the Ghrelin mRNA expression for Aviagen genotype lead to an elevation of muscle growth itself but its genetic potential did not respond enough according to the previous results (Table 4), recording finally higher (inefficient) FCR values. On the contrary, the significant negative regression of FCR ratio (-0.067) (Table 4) due to the excess of Ghrelin mRNA expression (lowest ΔCT mean) for Ross genotype may explain the decrease in its feed consumption, followed by a significant decrease in growth parameters. This finding confirms that Ghrelin is an anorixegenic hormone, that decreases appetite in Aviagen species and inhibits feeding behaviour [2,7,8,10-13,38-41] in chickens and as anorexigenic in all birds [15].
Conclusion
Under the experiment conditions, it can be concluded that even the five commercial hybrids that are recognized for only meat-type purpose with a similar growth rate performance, a difference in transcript level of Ghrelin mRNA among them was noticed. Gherlin hormone expression dose not correlated with growth parameters at early ages in broilers. At marketing age, the Ross hybrid had the highest hormone expression and it was associated with inhibition of its growth parameters. The Aviagen hybrid had the lowest hormone expression and this was associated with increase abdominal fat accumulation. The higher Gherlin hormone expression was responsible for decreasing muscles breast fat % for Arbor Acre broilers and muscles drumstick fat % in Hubbard broilers. The different transcription level of Ghrelin mRNA might be the result of different genetic origin of these hybrids, which could be explained by the existence of polymorphisms specific for their ancestor breeds that have not been identified so far. As a result, Ghrelin appears to correspond to growth-related quantitative traits differently, even among meat-type genotypes.
Acknowledgements
It would not have been possible to conduct this work without the advice and guides of Dr. Professor Farid Stino, Department of Animal production, Faculty of Agriculture, Cairo University in carrying out this research. I would like to express my sincere gratitude and appreciation to Assistant Professor Li Ma, Department of Animal and Avian Sciences, Maryland University, USA, for his information, encouragement and providing valued assistance.
References
- Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, et al. (1999) Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402: 656-660.
- Kaiya H, Van Der Geyten S, Kojima M, Hosoda H, Kitajima Y, et al. (2002) Chicken ghrelin: purification, cDNA cloning, and biological activity. Endocrinology 143: 3454-3463.
- Kaiya H, Miyazato M, Kangawa K, Peter RE, Unniappan S (2008) Ghrelin: a multifunctional hormone in non-mammalian vertebrates. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 149: 109-128.
- Korbonits M, Goldstone AP, Gueorguiev M, Grossman AB (2004) Ghrelin-a hormone with multiple functions. Frontiers in Neuroendocrinology 25: 27-68.
- Ueno H, Yamaguchi H, Kangawa K, Nakazato M (2005) Ghrelin: a gastric peptide that regulates food intake and energy homeostasis. Regul Pept 126: 11-19.
- Wada R, Sakata I, Kaiya H, Nakamura K, Hayashi Y, et al. (2003) Existence of ghrelin- munopositive and-expressing cells in the proventriculus of the hatching and adult chicken. Regul Pept 111: 123-128.
- Saito ES, Kaiya H, Tachibana T, Tomonaga S, Denbow DM, et al. (2005) Inhibitory effect of ghrelin on food intake is mediated by the corticotropin-releasing factor system in neonatal chicks. Regul Pept 125: 201-208.
- Saito ES, Kaiya H, Takagi T, Yamasaki I, Denbow DM, et al. (2002) Chicken ghrelin and growth hormone-releasing peptide-2 inhibit food intake of neonatal chicks. Eur J Pharmacol 453: 75-79.
- Ocłoń E, Pietras M (2011) Peripheral ghrelin inhibits feed intake through hypothalamo-pituitary-adrenal axis-dependent mechanism in chicken. J Anim Feed Sci 20: 118-130.
- Furuse M, Tachibana T, Ohgushi A, Ando R, Yoshimatsu T, et al. (2001) Intracerebroventricular injection of ghrelin and growth hormone releasing factor inhibits food intake in neonatal chicks. Neuroscience letters 301: 123-126.
- Geelissen SME, Swennen Q, Geyten SVD, Kuhn ER, Kaiya H, et al. (2006) Peripheral ghrelin reduces food intake and respiratory quotient in chicken. Domest Anim Endocrinol 30: 108-116.
- Kaiya H, Furuse M, Miyazato M, Kangawa K (2009) Current knowledge of the roles of ghrelin in regulating food intake and energy balance in birds. General and Comparative Endocrinology 163: 33-38.
- Shousha S, Nakahara K, Kojima M, Miyazato M, Hosoda H, et al. (2005) Different effects of peripheral and central ghrelin on regulation of food intake in the Japanese quail. General and Comparative Endocrinology 141: 178-183.
- Xu P, Siegel PB and Denbow DM (2011) Genetic selection for body weight in chickens has altered responses of the brain's AMPK system to food intake regulation effect of ghrelin, but not obestatin. Behavioural Brain Research 221: 216-226.
- Jonaidi H, Abbassi L, Yaghoobi MM, Kaiya H, Denbow DM, et al. (2012) The role of GABAergic system on the inhibitory effect of ghrelin on food intake in neonatal chicks. Neuroscience Letters 520: 82-86.
- Richards MP, Poch SM and McMurtry JP (2006) Characterization of turkey and chicken ghrelin genes, and regulation of ghrelin and ghrelin receptor mRNA levels in broiler chickens. General and Comparative Endocrinology 145: 298-310.
- Song Z, Liu L, Sheikhahmadi A, Jiao H, Lin H (2012) Effect of heat exposure on gene expression of feed intake Regul Pept in laying hens. BioMed Research International.
- Buyse J, Janssen S, Geelissen S, Swennen Q, Kaiya H, et al. (2009) Ghrelin modulates fatty acid synthase and related transcription factor mRNA levels in a tissue-specific manner in neonatal broiler chicks. Peptides 30: 1342-1347.
- Aghdam Shahryar H, Lotfi A (2016) Effects of peripheral administration of ghrelin antagonist [D-Lys 3]-GHRP-6 on growth performance and blood biochemical indices in broiler chickens. Archives Animal Breeding 59: 113-119.
- Goymann W, Lupi S, Kaiya H, Cardinale M, Fusani, L (2017) Ghrelin affects stopover decisions and food intake in a long-distance migrant. Proceedings of the National Academy of Sciences 201619565-201619565.
- Ghazanfari S, Tahmoorespur M, Nobari K (2010) Changes in ghrelin mRNA level, plasma growth hormone concentration and performance in different dietary energy and protein levels in broiler chicken. Italian Journal of Animal Science 9: e56-e56.
- Vizcarra JA, Wright H, Vizcarra A (2012) The effect of passive immunization against ghrelin on feed and water intake in turkeys. Poult Sci 91: 2305-2309.
- Seim I, Jeffery PL, Herington AC,
- Chopin LK (2015) Comparative analysis reveals loss of the appetite-regulating peptide hormone ghrelin in falcons. General and Comparative Endocrinology 216: 98-102.
- Takaya K, Ariyasu H, Kanamoto N, Iwakura H, Yoshimoto A, et al. (2000) Ghrelin strongly stimulates growth hormone release in humans. The Journal of Clinical Endocrinology & Metabolism 85: 4908-4911.
- Gondwe TNP & Wollny CBA (2005) Evaluation of the growth potential of local chickens in Malawi.
- Davis MJ (2010). “Contrast Coding in Multiple Regression Analysis: Strengths, Weaknesses, and Utility of Popular Coding Structures. Journal of Data Science 8: 61-73.
- Shavelson RJ (2016) Statistical reasoning for the Behavioral Sciences, translated by Güler N. Ankara: Pegema.
- Yakubu A and Ugbo SB (2011) An assessment of biodiversity in morphological traits of Muscovy ducks in Nigeria using discriminant analysis. Proceedings of 2010 International Conference on Biology Environment and Chemistry, IACSIT Press, Singapore, pp. 389-391
- Fang M, Nie Q, Luo C, Zhang D, Zhang X (2007) An 8bp indel in exon 1 of Ghrelin gene associated with chicken growth. Domestic Animal Endocrinology 32: 216-225.
- Fang M, Nie Q, Luo C, Zhang D, Zhang X (2010) Associations of GHSR gene polymorphisms with chicken growth and carcass traits. Molecular Biology Reports 37: 423-428.
- Kgwatalala PM, Thutwa K, Nsoso SJ (2012) Single nucleotide polymorphisms in ghrelin gene and the resulting genetic variants at ghrelin locus in different strains of indigenous Tswana chickens. African Journal of Biotechnology 11: 10534-10540.
- Li CC, Li K, Li J, Mo DL, Xu RF, Chen, et al. (2006) Polymorphism of ghrelin gene in twelve Chinese indigenous chicken breeds and its relationship with chicken growth traits. ASIAN AUSTRALASIAN Journal of Animal Sciences 19: 153-153.
- Huang N, Cogburn LA, Agarwal SK, Marks HL, Burnside J (1993) Overexpression of a truncated growth hormone receptor in the sex-linked dwarf chicken: evidence for a splice mutation. Molecular Endocrinology 7: 1391-1398.
- Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, et al. (2003) A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425: 832-836.
- Lei M, Luo C, Peng X, Fang M, Nie Q, et al. (2007) Polymorphism of growth-correlated genes associated with fatness and muscle fiber traits in chickens. Poult Sci 86: 835-842.
- Nie Q, Fang M, Xie L, Peng X, Xu H, et al. (2010) Molecular characterization of the ghrelin and ghrelin receptor genes and effects on fat deposition in chicken and duck. BioMed Research International.
- Ropka-Molik K, Oczkowicz M, Piórkowska K, Różycki M, Romanek J, et al. (2011) New polymorphisms and expression of the porcine ghrelin (GHRL) gene in different pig breeds. Journal of Animal and Feed Sciences 663: 127-129.
- Xu QL, Tang GW, Zhang QL, Huang YK, Liu YX, et al. (2011) The FABP4 gene polymorphism is associated with meat tenderness in three Chinese native sheep breeds, pp. 1-6.
- Abdi H, Williams L (2010) Contrast Analysis: In Encyclopedia of Research Design, edited by Salkind NJ, Dougherty DM, Frey B Thousand Oaks: Sage 243-251.
- Nie Q, Lei M, Ouyang J, Zeng H, Yang G, et al. (2005) Identification and characterization of single nucleotide polymorphisms in 12 chicken growth-correlated genes by denaturing high performance liquid chromatography. Genetics Selection Evolution 37: 339-341.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences