ISSN : 2573-4466
Insights in Enzyme Research
Cloning, Overexpression and Characterization of a Catalase from a Marine Acinetobacter Bacterium
Wei Wang1,2,3, Xinhua Fu4, Jingjing Sun1,2,3, Junzhong Liu1,2,3, Jianhua Hao1,2,3* and Mi Sun1,2,3*
1Key Laboratory of Sustainable Development of Polar Fishery, Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, PR China
2Laboratory for Marine Drugs and Bioproducts, Qingdao National Laboratory for Marine Science and Technology, Qingdao, PR China
3Jiangsu Collaborative Innovation Center for Exploitation and Utilization of Marine Biological Resource, Lianyungang 222005, China
4Weifang Medical University, Weifang, PR China
- *Corresponding Author:
- Jianhua Hao
Key Laboratory of Sustainable Development of Polar Fishery
Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute
Chinese Academy of Fishery Sciences, Qingdao, PR China
Tel: +86-532-85841193
E-mail: haojh@ysfri.ac.cn
Mi Sun
Key Laboratory of Sustainable Development of Polar Fishery
Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute
Chinese Academy of Fishery Sciences, Qingdao, PR China
Tel: +86-532-85819525
E-mail: sunmi@ysfri.ac.cn
Received Date: December 07, 2018; Accepted Date: December 31, 2018; Published Date: January 10, 2019
Citation: Wang W, Fu X, Sun J, Liu J, Hao J, et al. (2019) Cloning, Overexpression and Characterization of a Catalase from a Marine Acinetobacter Bacterium. Insights Enzyme Res. 2019, Vol.3 No.1: 1.
Copyright: © 2019 Wang W, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Objective: The marine catalase YS0810CAT gene was cloned and overexpressed from Acinetobacter sp. YS0810, and high stability of the recombinant enzyme was validated, which made it important for potential applications in the elimination of hydrogen peroxide from industrial process-generated streams.
Methods: The gene was cloned by PCR and overexpressed in Escherichia coli. Evolutionary analyses of this enzyme were conducted with the MEGA software. Anion exchange was applied to purify the recombinant enzyme. The effects of pH and temperature on the activity and stability of YS0810CAT were measured.
Results: The gene consists of 1,518 bp and belongs to Clade 3 of monofunctional catalases. The maximum protein production was obtained with 0.8 mM IPTG, a post-induction temperature of 37°C, and a post-induction time of 8 h. The recombinant protein was most active at 60°C and pH 11.
Conclusion: The effects of pH and temperature on the activity and stability of the wild type and recombinant YS0810CAT are similar. The protocol for the preparation of recombinant YS0810CAT could aid enzyme crystallization; moreover, improvement in its properties may be possible through protein engineering.
Keywords
Monofunctional catalase; Gene cloning; Enzyme expression; Marine Acinetobacter; Thermostability; Alkali stability; Activity
Introduction
Hydrogen peroxide, as an inevitable by-product of aerobic metabolism, can damage nucleic acids, proteins, or lipids in cells or tissues [1]. Catalases (EC 1.11.1.6) are essential elements in the cellular defense mechanism against oxidative stress, as they are responsible for the catalysis of excess hydrogen peroxide to water and oxygen.
Generally, catalases can be divided into three main groups: monofunctional catalases, catalase-peroxidases and manganese (Mn) catalase. Monofunctional catalases and catalase-peroxidases are heme-containing enzymes and are ubiquitously expressed in most prokaryotic and eukaryotic microorganisms. The well-studied monofunctional catalases generally contain four identical subunits and do not exhibit peroxidase activity [1]; their gene family could be phylogenetically divided into three groups or clades [1,2]. Clade 1 and Clade 3 contain catalases composed of smallsubunits, while Clade 2 contains large subunit catalases. Most monofunctional catalases are grouped in Clade 3 [2].
As usage of hydrogen peroxide in industrial settings grows, catalases have a potential application in the degradation of hydrogen peroxide derived from process streams in the textile or food industry. This is extremely important since hydrogen peroxide can interfere with subsequent steps within the industrial process [3]. In a previous study, a highly active catalase YS0810CAT from the marine Gammaproteobacteria Acinetobacter sp. YS0810 was purified and characterized as monofunctional catalase, with high alkali-stability and thermostability [4]. Possible applications of YS0810CAT were found in a wide range of industrial processes. Here, we describe the methodology for cloning and expressing YS0810CAT with the purpose of providing the basis for future crystallization and protein engineering.
Materials and Methods
Bacterial strains and cultivation
The Acinetobacter sp. YS0810 strain (CCTCC No. M2011067) was grown in a 28°C incubator with constant agitation at 220 rpm for 24 h. The medium composition was 1% peptone, 1% beef extract and 0.5% NaCl (pH 7.2). Escherichia coli DH5α and BL21 (DE3) strains were routinely grown in Luria Bertani (LB) medium, supplemented with the relevant antibiotics (100 μg/mL Ampicillin or 25 μg/mL Kanamycin), at 37°C at 200 rpm.
DNA manipulation in-vitro
Routine DNA manipulation was performed as described in Current Protocols in Molecular Biology [5]. All enzymes were purchased from Takara (Dalian, Liaoning, China). Oligonucleotides and other routinely-used chemicals were purchased from Sangon (Shanghai, China). DNA sequence analyses were also performed by Sangon.
Gene cloning and expression
The N-terminal sequence (SQDPKKCPVTHLTTE) of YS0810CAT [4] was submitted to a BLASTP search on the NCBI and the catalase (WP_043041729.1, KatE) from A. baumannii NBRC 110492 was found to be a positive match (i.e., to have an identical sequence). PCR primers FF (5’- ATGAGTCAAGACCCTAAAAAATG-3’) and FR (5’- AAGAAAACTTGGTAAACCTTTAG-3’) for cloning the YS0810CAT gene were designed according to the 5’ and 3’ end sequence of the A. baumannii catalase gene (NZ_BBTD01000103: 196-1716). 50 μl PCR mixture contained 1 μl of template, 25 μl of Premix Ex Taq PCR master mix (Takara), 2 μl of each primer (10 μM), and 20 μl of Milli-Q water. The PCR conditions were: 30 cycles of 94°C for 60 seconds, 48°C for 60 seconds and 72°C for 60 seconds.
The denaturation time of the first cycle was prolonged to 4 minutes, and the extension time of the last cycle was increased to 7 minutes. The PCR product was purified using an EZ Spin Column DNA Gel Extraction Kit (Sangon), ligated with the pEASY-T1 T-vector (TransGen, Beijing, China), and cloned in E. coli DH5α competent cells (TransGen). Three clones were randomly picked and sequenced using the vector primers M13-20 (5’-GTAAAACGACGGCCAG-3’) and M13-27 (5’- GGAAACAGCTATGACCATG-3’). The sequences of the inserted DNA fragment in the T-vector were analyzed and determined as the amplified gene encoding YS0810CAT. Then the YS0810CAT gene was cloned into the pET-28a expression vector using primers containing BamHI and XhoI restriction sites (underlined), respectively: F2F (5’- aaaGGATCCATGAGTCAAGACCCTAAAAAATG-3’) and F2R (5’- aaaCTCGAGAAGAAAACTTGGTAAACCTTTAGC-3’), E. coli DH5α was also used as the host.
The resulting plasmid, pET28CAT, was then transformed into E. coli BL21 (DE3) using the expression protocol. DNA fragments were sequenced by the ThermoFisher 3730xl DNA Analyzer in Sangon (Shanghai, China). The sequences of the YS0810CAT gene and the corresponding protein have been submitted to the Genbank database (Accession No. KJ612073 and AHY00946). Catalase sequence alignments were performed using the PROMALS3D website [6]. Evolutionary analyses were conducted with the MEGA software version 7 [7].
Characterization of the recombinant catalase
E. coli BL21 (DE3) cells containing pET28CAT were harvested by centrifugation (10,000 g, 10 minutes), re-suspended in phosphate buffer (20 mM, pH 7.5), and disrupted by ultrasonication. The supernatant was obtained by centrifugation at 10,000 g, for 10 minutes, at 4°C and was applied to a HiPrep DEAE FF 16/10 column (GE Healthcare Life Sciences) equilibrated with buffer A (50 mM pH 7.5 Tris·Cl). Proteins were eluted with a linear gradient of NaCl from 0 to 0.3 M in the buffer A with a flow rate of 1.5 mL/min. The absorbance of the effluent stream at 280 nm and 405 nm were recorded and fractions corresponding to the peaks of 405 nm, containing most catalase activity, were pooled and concentrated by Amicon® Ultra 15 mL centrifugal filter devices (Merck Millipore). The chromatography was done by an ÄKTA Explorer 10S Protein Purification Systems (GE Healthcare Life Sciences).
The target protein was identified by SDS-PAGE [8] and the catalase activity (i.e., elimination of H2O2) was measured spectrophotometrically by monitoring the decrease in absorbance at 240 nm [9]. Peroxidase-like activity was determined spectrophotometrically at 450 nm in 50 mM citric acid buffer (pH 4.5) containing 0.3 mM O-phenylenediamine [10]. The supernatant containing the wild type enzyme was also prepared as previously described [4].
To determine the YS0810CAT pH stability, the residual activity was measured after incubation of the enzyme in phosphate solutions buffered at different pH, at 4°C for 3 hours. The thermostability of YS0810CAT was determined after the enzyme was incubated for 30 minutes in 50 mM phosphate buffer (pH 7.0) at various temperatures.
Results
Gene cloning and sequence analysis
The YS0810CAT gene (1518 bp) encodes a protein of 506 amino acids. The predicted molecular mass of the monomer was 57,275 Daltons and was similar to that reported earlier [4]. The YS0810CAT gene was successfully amplified by PCR. The amplified YS0810CAT sequence showed an identity value of 505/506 (99%) with the A. baumannii catalase.
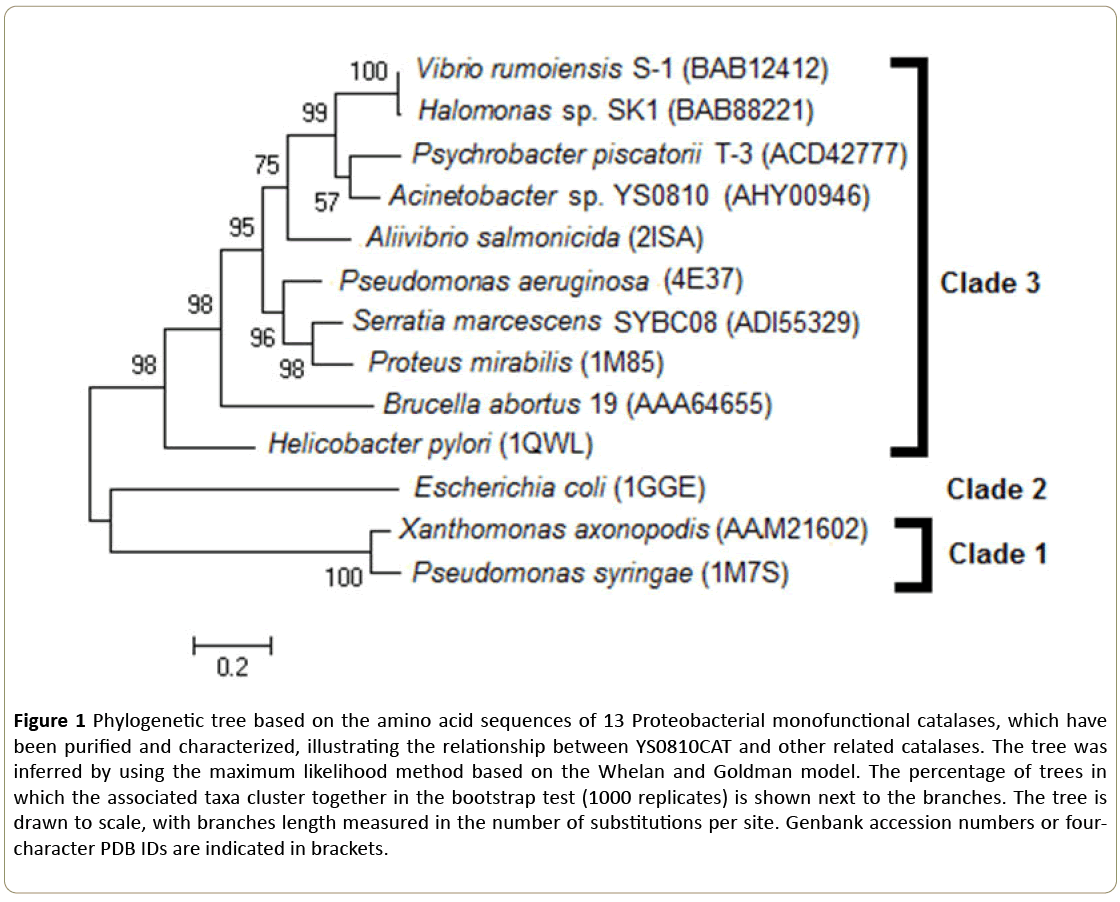
The phylogenetic tree of YS0810CAT and other Proteobacterial catalases (Figure 1) shows that YS0810CAT belongs to the Clade 3 of the monofunctional catalases, in accordance with the previous conclusion that the YS0810CAT is a small-subunit monofunctional catalase [4]. The Clade of which YS0810CAT belongs to could be only determined by phylogenetic analysis rather than the biochemical properties of the enzyme.
Figure 1: Phylogenetic tree based on the amino acid sequences of 13 Proteobacterial monofunctional catalases, which have been purified and characterized, illustrating the relationship between YS0810CAT and other related catalases. The tree was inferred by using the maximum likelihood method based on the Whelan and Goldman model. The percentage of trees in which the associated taxa cluster together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branches length measured in the number of substitutions per site. Genbank accession numbers or fourcharacter PDB IDs are indicated in brackets.
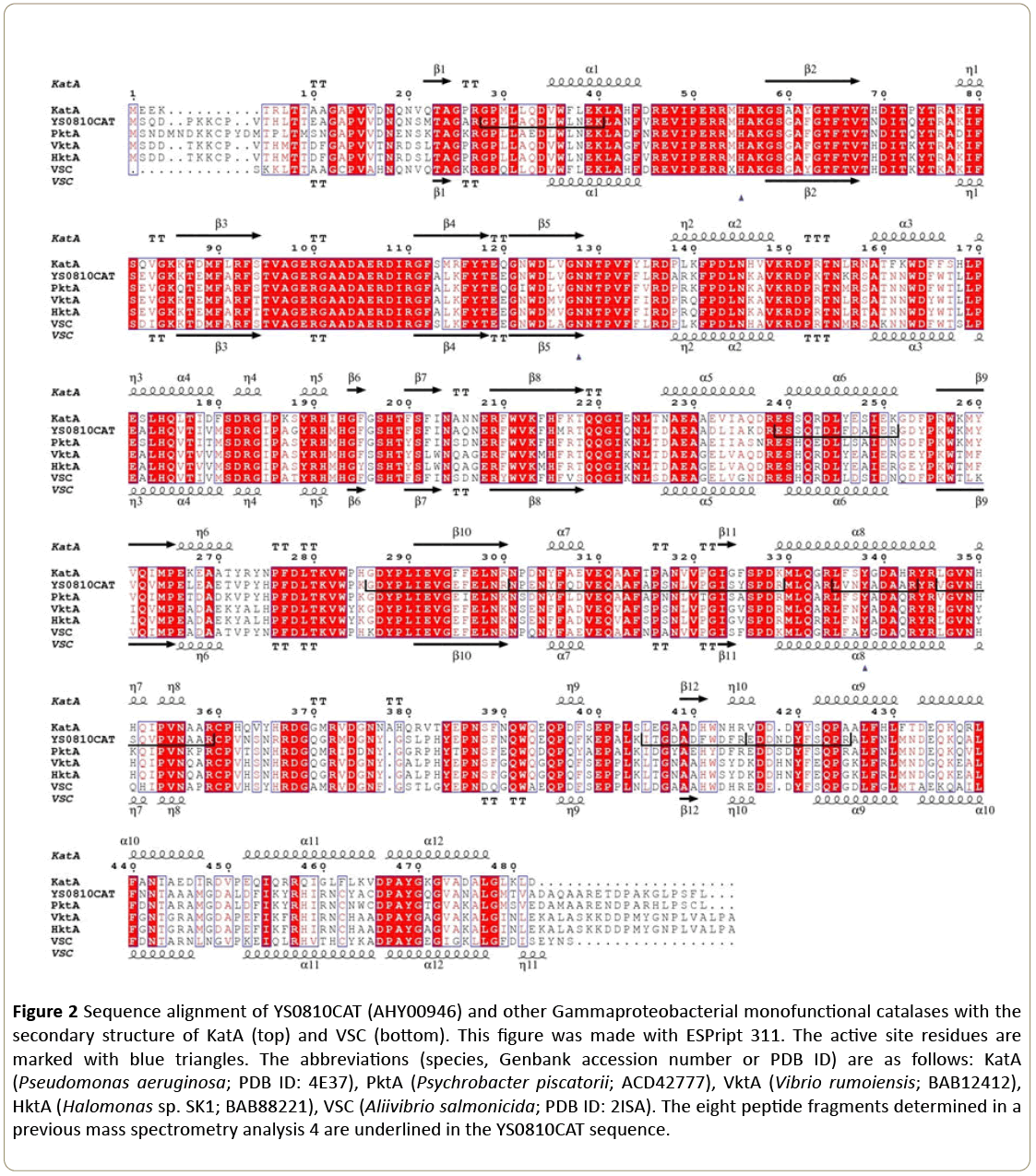
In a previous work, eight peptide fragments belonging to the protein sequence of YS0810CAT were obtained with MALDI-TOF/TOF tandem mass spectrometry [4]. The cloned gene also showed the presence of these peptide fragments in the catalase (Figure 2), which indicated that this was indeed the YS0810CAT gene.
Figure 2: Sequence alignment of YS0810CAT (AHY00946) and other Gammaproteobacterial monofunctional catalases with the secondary structure of KatA (top) and VSC (bottom). This figure was made with ESPript 311. The active site residues are marked with blue triangles. The abbreviations (species, Genbank accession number or PDB ID) are as follows: KatA (Pseudomonas aeruginosa; PDB ID: 4E37), PktA (Psychrobacter piscatorii; ACD42777), VktA (Vibrio rumoiensis; BAB12412), HktA (Halomonas sp. SK1; BAB88221), VSC (Aliivibrio salmonicida; PDB ID: 2ISA). The eight peptide fragments determined in a previous mass spectrometry analysis 4 are underlined in the YS0810CAT sequence.
In order to understand the residues responsible for the observed alkali-stability and thermostability features of YS0810CAT, the sequence of YS0810CAT was aligned and compared to that of other Gammaproteobacterial catalases (Figure 2). The sequence identity at the C-terminus (residues 350-480) is considerably lower than in the other domains.
The secondary structure of YS0810CAT, however, might resemble those of KatA and VSC because of the overall high structural conservation of monofunctional catalase. The highly conserved catalytic residues (H61, N134, and Y344) of YS0810CAT are indeed the same as in the other monofunctional catalases [2], suggesting that YS0810CAT has the same catalytic mechanism as the catalases with experimentally-determined structures.
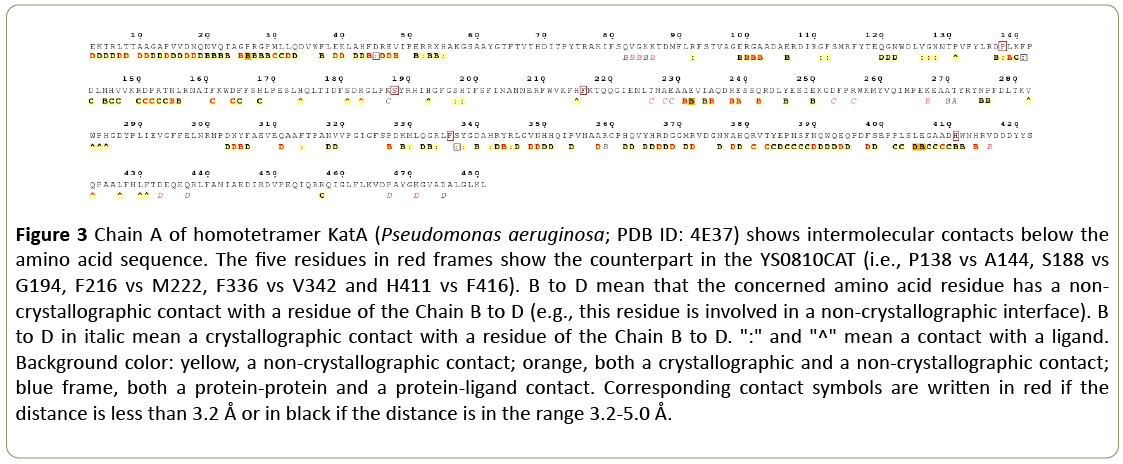
Figure 2 also reveals several mutations that are unique to the YS0810 strain. These mutations include A144 (P138 in other catalases based on alignment), G194 (S188 or T188 in others), M222 (F216 in others), V342 (F336 in others) and F416 (H411 in others). The possible location of the five mutations in the YS0810CAT tertiary structure might be inferred from the KatA catalase structure, using ENDscript 2 software [11] (Figure 3).
Figure 3: Chain A of homotetramer KatA (Pseudomonas aeruginosa; PDB ID: 4E37) shows intermolecular contacts below the amino acid sequence. The five residues in red frames show the counterpart in the YS0810CAT (i.e., P138 vs A144, S188 vs G194, F216 vs M222, F336 vs V342 and H411 vs F416). B to D mean that the concerned amino acid residue has a noncrystallographic contact with a residue of the Chain B to D (e.g., this residue is involved in a non-crystallographic interface). B to D in italic mean a crystallographic contact with a residue of the Chain B to D. ":" and "^" mean a contact with a ligand. Background color: yellow, a non-crystallographic contact; orange, both a crystallographic and a non-crystallographic contact; blue frame, both a protein-protein and a protein-ligand contact. Corresponding contact symbols are written in red if the distance is less than 3.2 Å or in black if the distance is in the range 3.2-5.0 Å.
These residues could be involved in protein-ligand interactions (A144, M222 and V342), or be located at the interface between catalase subunits (G194, V342 and F416), which might confer enhanced stability and important kinetic properties.
Catalase expression and characterization
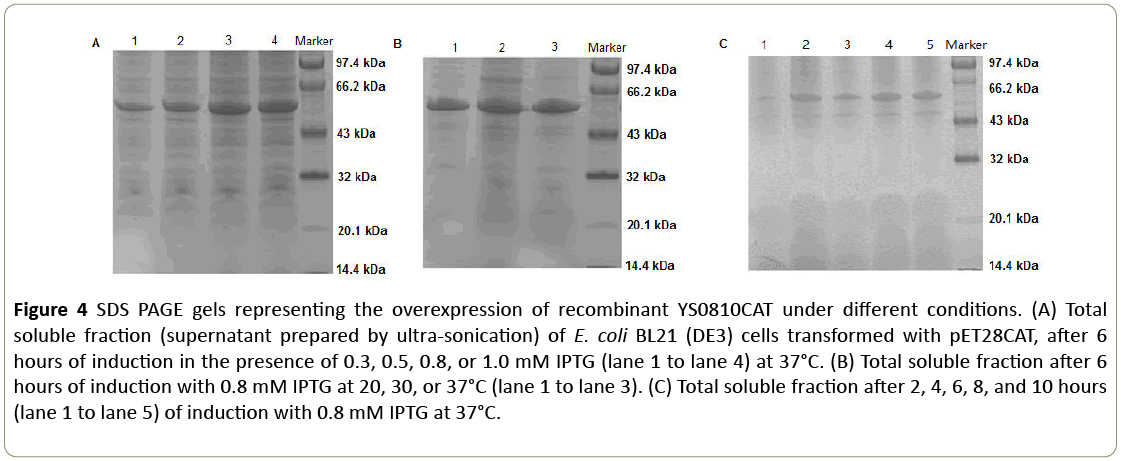
E. coli BL21 (DE3) cells, transformed with the pET28CAT plasmid, were grown at mid-log phase (OD600=0.6) before the expression of the protein was induced with isopropyl β-D-1- thiogalactopyranoside (IPTG). The recombinant YS0810CAT was highly expressed in the soluble fraction. To optimize the production of soluble YS0810CAT in E. coli , different inductive conditions (i.e., IPTG concentrations, post-induction temperature, and time) were tested.
As illustrated in Figure 4, the maximum protein production was obtained with 0.8 mM IPTG, a post-induction temperature of 37°C, and a post-induction time of 8 hours.
Figure 4: SDS PAGE gels representing the overexpression of recombinant YS0810CAT under different conditions. (A) Total soluble fraction (supernatant prepared by ultra-sonication) of E. coli BL21 (DE3) cells transformed with pET28CAT, after 6 hours of induction in the presence of 0.3, 0.5, 0.8, or 1.0 mM IPTG (lane 1 to lane 4) at 37°C. (B) Total soluble fraction after 6 hours of induction with 0.8 mM IPTG at 20, 30, or 37°C (lane 1 to lane 3). (C) Total soluble fraction after 2, 4, 6, 8, and 10 hours (lane 1 to lane 5) of induction with 0.8 mM IPTG at 37°C.
Initial purification experiments were carried out according to the His-tag chromatographic method, but the recombinant protein did not bind to the HisTrap FF column. The terminal His-tags might be buried in the structure and thus not prone to bind to the Ni2+ resins [12]. An alternative anion exchange was thus applied as described in the aforementioned methods.
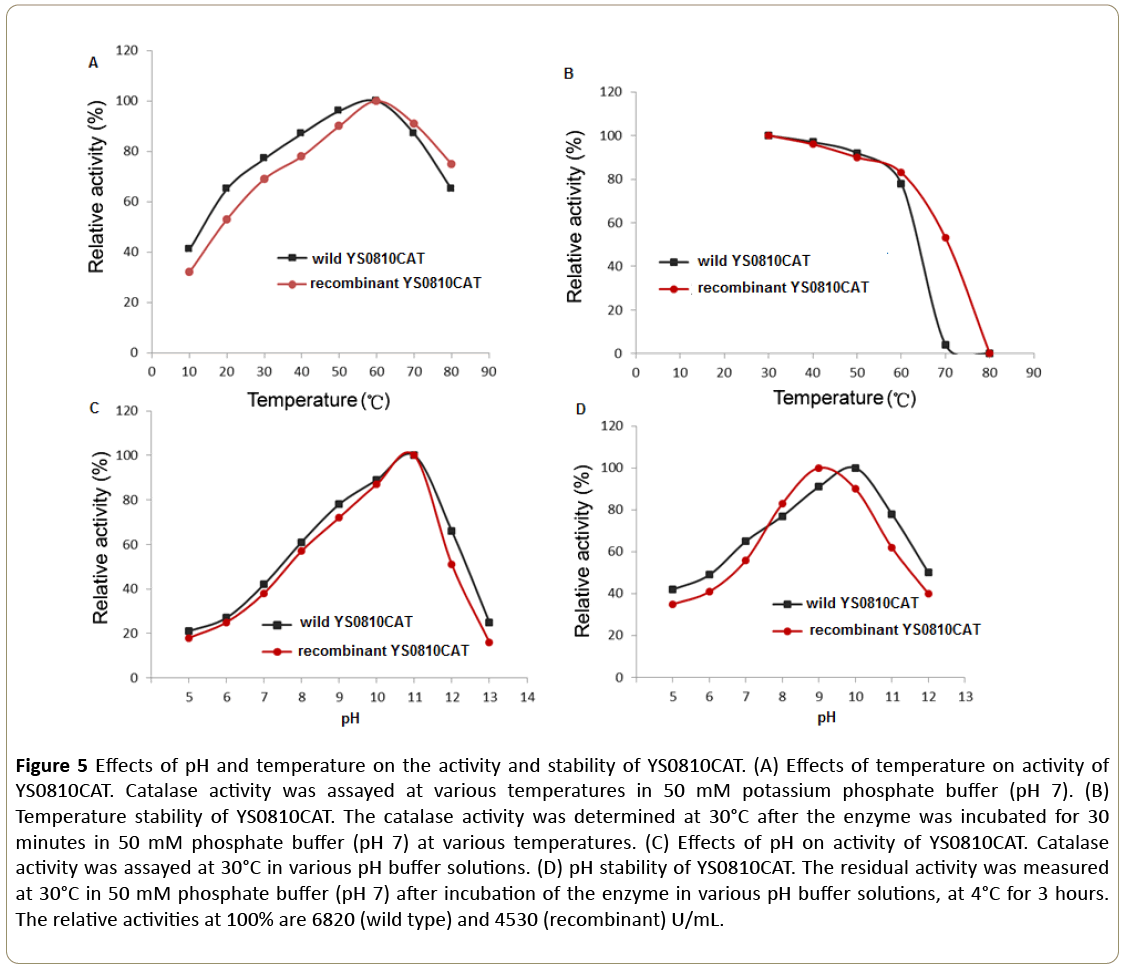
Peroxidase like activity was not found in this recombinant catalase, which demonstrated the purified catalase did not contain the catalase-peroxidase HPI from E. coli [13]. Both the wild type and recombinant YS0810CAT exhibited similar temperature and pH dependent activities (Figure 5A and 5C). The YS0810CAT was most active at 60°C and pH 11.0 in 50 mM phosphate buffer. There were no marked differences in thermostability between the wild type and recombinant YS0810CAT despite a sharp decrease in the activity of the wild type at temperatures above 60°C (Figure 5B). The enzyme was stable between pH 8.0-10.0, and the YS0810CAT wild type retained more than 70% of the original activity after 3 hour incubation at pH 11.0 (Figure 5D). In conclusion, the effects of pH and temperature on the activity and stability of two types of YS0810CAT were mostly similar, and the differences might be due to the lack of purity in both types of YS0810CAT.
Figure 5: Effects of pH and temperature on the activity and stability of YS0810CAT. (A) Effects of temperature on activity of YS0810CAT. Catalase activity was assayed at various temperatures in 50 mM potassium phosphate buffer (pH 7). (B) Temperature stability of YS0810CAT. The catalase activity was determined at 30°C after the enzyme was incubated for 30 minutes in 50 mM phosphate buffer (pH 7) at various temperatures. (C) Effects of pH on activity of YS0810CAT. Catalase activity was assayed at 30°C in various pH buffer solutions. (D) pH stability of YS0810CAT. The residual activity was measured at 30°C in 50 mM phosphate buffer (pH 7) after incubation of the enzyme in various pH buffer solutions, at 4°C for 3 hours. The relative activities at 100% are 6820 (wild type) and 4530 (recombinant) U/mL.
Discussion
Thousands of whole genomes belonging to the Acinetobacter strains are present in Genbank. For that reason, the result of the BLASTP search of YS0810CAT (AHY00946) included numerous Acinetobacter catalases sharing a high identity value ( ≥ 95%) with the target sequence. Those Acinetobacter catalases, automatically annotated from the genome sequences, have not yet been purified and characterized. Therefore, our study might also provide help in deducing the properties of those enzymes. For example, the capacity of A. baumannii to degrade H2O2 is largely dependent on katE [14]. Although the biochemical characteristics of the katE were not studied, it could be inferred from that of YS0810CAT, because katE showed an identity value of 99% with YS0810CAT.
Among the cloned and purified catalases, YS0810CAT had the highest identity (82%) to PktA (ACD42777) from Psychrobacter piscatorii T-3 [15]. YS0810CAT was also aligned with the catalases in the protein data bank database, where the highest identity (71%) was with the KatA (PDB ID 4E37) catalase of Pseudomonas aeruginosa. In bacterial taxonomy, Acinetobacter, Psychrobacter, and Pseudomonas belong to Pseudomonadales in Gammaproteobacteria, explaining the high identity between YS0810CAT and PktA and KatA.
Although the amino acid sequence of YS0810CAT showed the highest similarity with the PktA catalase, among the others, their biochemical characteristics are very difference. PktA did not exhibit a clear temperature dependence between 10 and 55°C [15], while, in the same range, YS0810CAT exhibited a clear temperature dependence profile (Figure 5A). Moreover, PktA has a broad optimum pH (pH 5.0 to 10.0), while YS0810CAT activity is higher between pH 10.0 to 11.0. Recently, several Acinetobacter strains have been reported to express catalases with relatively high specific activities, which also related to their extremotolerance. For example, the catalase from A. gyllenbergii 2P01AA is pH stable, alkalitolerant, and halotolerant [16], while the high activity catalase expressed by the Acinetobacter strain Ver7 played an important role in UV tolerance [17]. Although the gene of the A. gyllenbergii 2P01AA catalase was not cloned, most of the amino acid residues of the catalase were identified by mass spectrometry [16]; the amino acid sequence showed high similarity (87%) to the sequence of the YS0810CAT, which could explain the similar stability of the two Acinetobacter catalases. Like the previously reported catalase from A. gyllenbergii 2P01AA, YS0810CAT also had an alkali pH optimum and displayed alkali-tolerance. The recombinant YS0810CAT catalase retained 83% of the activity after 30 min incubation at 60°C. To our knowledge, this is the most thermostable monofunctional catalase of the Clade 3 among reported mesophilic bacteria. The high activity and stability of the abovementioned catalases suggest that the Acinetobacter genus might be of importance for discovering novel antioxidant enzymes for potential biocatalysts.
The preparation of recombinant YS0810CAT may lay the foundation for elucidating the protein structure, which is critical to understand the high thermostability and alkalistability of this enzyme. The potential of protein engineering for the enhancement of the catalytic properties of YS0810CAT is also dependent on successful gene cloning and protein expression. The theoretical and applied research on YS0810CAT will focus on the peculiar biochemical characteristics typical of this enzyme activity and stability.
Acknowledgments
This study was supported by The Scientific and Technological Innovation Project Financially Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2016ASKJ14); Central Public-interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022018006) and The Financial Fund of the Ministry of Agriculture and Rural Affairs, PR China (No. NFZX2013).
Conflict of Interest
The authors declare no conflict of interest.
References
- Klotz MG, Loewen PC (2003) The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from Bacteria into Eukaryota. Mol Biol Evol 20: 1098-1112.
- Zamocky M, Furtmüller PG, Obinger C (2008) Evolution of catalases from Bacteria to humans. Antioxid Redox Sign 10: 1527-1547.
- Sooch BS, Kauldhar BS, Puri M (2014) Recent insights into microbial catalases: Isolation, production and purification. Biotechnol Adv 32: 1429-1447.
- Fu X, Wang W, Hao J, Zhu X, Sun M (2014) Purification and characterization of catalase from marine bacterium Acinetobacter sp. YS0810. Biomed Research International.
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al. (2003) Current Protocols in Molecular Biology. John Wiley & Sons, Hoboken.
- Pei J, Tang M, Grishin NV (2008) PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res 36: 30-34.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Wang W, Sun M, Liu W, Zhang B (2008) Puriï¬ÂÂcation and characterization of a psychrophilic catalase from Antarctic Bacillus. Can J Microbiol 54: 823-828.
- Hamilton TM, Dobie-Galuska AA, Wietstock SM (1999) The O-phenylenediamine-horseradish peroxidase system: enzyme kinetics in the general chemistry laboratory. J Chem Educ 76: 642.
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: 320-324.
- Lorentzen MS, Moe E, Jouve HM, Willassen NP (2006) Cold adapted features of Vibrio salmonicida catalase: characterisation and comparison to the mesophilic counterpart from Proteus mirabilis. Extremophiles 10: 427-440.
- Claiborne A, Fridovich I (1979) Purification of the O-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J Biol Chem 254: 4245-4252.
- Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, et al. (2016) KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148: 31-40.
- Kimoto H, Yoshimune K, Matsuyma H, Yumoto I (2012) Characterization of catalase from psychrotolerant Psychrobacter piscatorii T-3 exhibiting high catalase activity. Int J Mol Sci 13: 1733-1746.
- Muster N, Derecho I, Dallal F, Alvarez R, McCoy KB, et al. (2015) Purification, biochemical characterization, and implications of an alkali-tolerant catalase from the spacecraft-associated and oxidation-resistant Acinetobacter gyllenbergii 2P01AA. Astrobiology 15: 291-300.
- Di Capua C, Bortolotti A, Farías ME, Cortez N (2011) UV-resistant Acinetobacter sp. isolates from Andean wetlands display high catalase activity. FEMS Microbiol Lett 317: 181-189.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences