
Page 20
Volume 05
Journal of Infectious Diseases and Treatment
ISSN: 2472-1093
JOINT EVENT
Applied Microbiology-2019 & Antibiotics 2019
Immunology 2019
October 21-22, 2019
October 21-22, 2019 Rome, Italy
&
&
8
th
Edition of International Conference on
Antibiotics, Antimicrobials & Resistance
12
th
International Conference on
Allergy & Immunology
6
th
World Congress and Expo on
Applied Microbiology
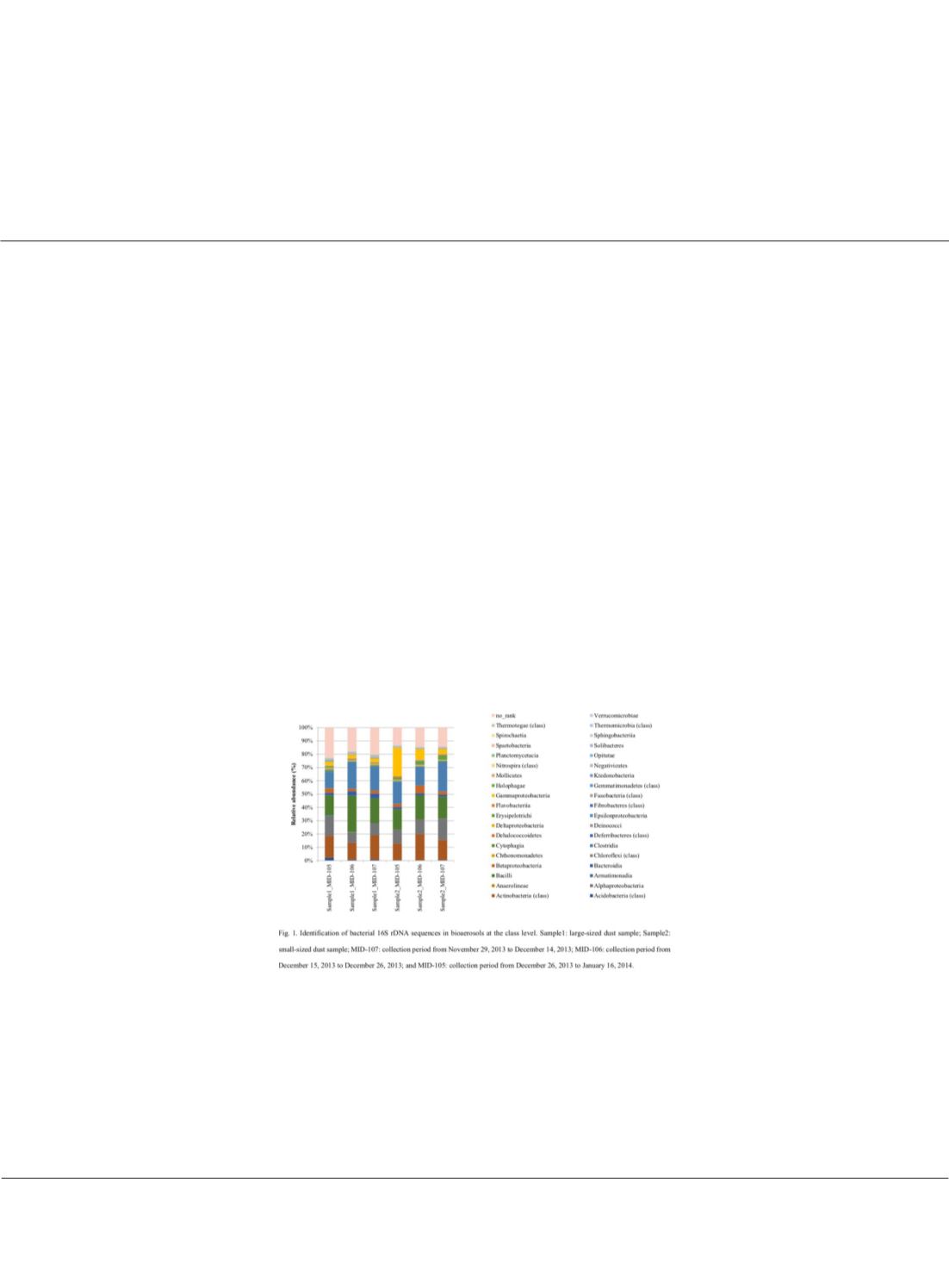
Identification of bioaerosols from environmental samples in the AIST, Tsukuba, Japan
Panyapon Pumkaeo, Wenhao Lu, Youki Endou, Tomohiro Mizuno, Junko Takahashi
and
Hitoshi Iwahashi
Gifu University, Japan
T
he bioaerosols are the atmosphere particles, mists or dust of µm range, associated with metabolically active or
inactive viable particles. They contain living organism’s included microorganisms such as viruses, bacteria, and
fungi also plant material as well as pollen. Next Generation Sequencing (NGS) is a novel method of DNA sequencing
that quickly and efficiently read the underlying sequence of an organism by means of massively parallel sequencing.
The aim of this study is identifying organisms which contained in environmental samples by using NGS. This study
monitored the environmental sample (bioaerosols) from November 2013 to January 2015 for 50 days using air
samples were collected at AIST, Tsukuba, Japan. Samples were bio-analyzed using a next-generation sequencing
method. In this study, we used two NGS platform, GS FLX+ (Roche 454 sequencing) and Illumina Misep. The sample
was detected plants, eukaryotes and bacteria. The sample was divided into two subgroup subgroups according to the
size of its bioaerosols, large subgroup contains bioaerosols whose diameter is bigger than 3.3μm, and small subgroup
contains those smaller than 3.3μm.The most abundant bacteria in several samples were of the Actinobacteria (class),
Alphaproteobacteria, Bacilli and Clostridia. For the animal detection using internal transcribed spacer 1, only
uncultured fungi were detected in more than half of the hits, with a high number of Cladosporium sp. in the samples.
For the plant identification, the ITS1 information only matched fungal species. However, targeting of the rbcL region
revealed diverse plant information, such as
Medicago papillosa
. In conclusion, traces of bacteria, fungi, and plants
could be detected in the bioaerosols, but not of animals using those primers.
Recent Publications
1.
Choi, Jae-Hoon, Ayaka Kikuchi, Panyapon Pumkaeo, Hirofumi Hirai, Shinji Tokuyama and Hirokazu
Kawagishi (2016) Bioconversion of AHX to AOH by resting cells of Burkholderia contaminans CH-1.
Bioscience, Biotechnology and Biochemistry 80 (10):2045-2050.
2.
Panyapon Pumkaeo. 2018. "Identification of Bacteria from bioaerosl at AIST, Tsukuba, Japan." Proceedings
of International Symposium on Animal Production and Consevation for Sustainable Development 2018
UGSAS-GU & BWEL Joint Poster Session on Agricultureal and Basin Water Environmental Sciences
2018:P18.
Panyapon Pumkaeo et al., J Infec Dis Treat 2019, Volume: 05
















