Transcriptional Regulation of Ribosomal Protein Genes in Yeast and Metazoan Cells
Sandra Moreira-Ramos1, Fabiola Urbina1, Edio Maldonado1* and Diego A Rojas2
1Deparment of Biología Celular and Molecular, Instituto de Ciencias Biomédicas, Universidad de Chile, Santiago,Chile
2Facultad de Medicina, Deparment of Microbiology, Instituto de Ciencias Biomédicas, Universidad de Chile, Chile
- *Corresponding Author:
- Edio Maldonado
Deparment of Biología Celular and Molecular
Instituto de Ciencias Biomédicas
Universidad de Chile, Santiago, Chile
Tel: 56-2-29786207
E-mail: emaldona@med.uchile.cl
Received: July 24, 2018; Accepted: July 28, 2018; Published: August 06, 2018
Citation: Moreira-Ramos S, Urbina F, Maldonado E, Rojas DA (2018) Transcriptional Regulation of Ribosomal Protein Genes in Yeast and Metazoan Cells. J Mol Cell Biochem Vol.2 No.1:5.
Abstract
Several cellular processes are regulated through coordinated mechanisms including those related to the expression of gene regulatory networks or transcriptional modules. The identification of these transcriptional modules is essential to understand how different genes are coordinately expressed in response to both positive or negative stimulus. In eukaryotic cells, due to the large number of genes, the identification of transcriptional modules is difficult. However, during the last years several studies have been performed to understand the coordinated expression of ribosomal protein genes (RPGs). Those genes form a gene regulatory network and contain in their promoter sequences one or more common cis DNA elements which can control and regulate their expression. Such is the case of the yeast Saccharomyces cerevisiae, where the conserved Rap1 sequence is the key element in the recruiting of the RNA polymerase II transcriptional machinery. This element binds the transcription factor Rap1. However, Rap1 sequence is absent in the yeast Schizosaccharomyces pombe, where this element was replaced by the HomolD-box sequence, which binds the transcription factor Rrn7. Interestingly, neither of both elements have been identified in metazoan RPGs promoters. However, analyses in Drosophila have shown the presence of the TCT element in the RPGs promoters, which may be recognized by the factor M1PB. The study of each of these elements and the transcription factors which are able to bind them, is essential to understand the coordination of the expression of RPGs, which are fundamental in ribosome biogenesis and in the cellular response to environmental cues.
Keywords
Ribosomal protein genes; RPGs promoters; Gene transcription
Introduction
Perhaps, the construction and understanding of gene regulatory networks (transcriptional modules) is one of the most important and difficult task to perform. However, it is necessary to decipher how different genes in an organism can be regulated and coordinately expressed in response to both positive and negative signals. Since the eukaryotic cells contain several thousands of genes, the identification of such transcriptional modules is a challenging event.
The ribosome is the protein-synthesizing organella in all living cells and in the yeast Saccharomyces cerevisiae is composed for 4 rRNA and 79 ribosomal proteins (RP) encoded by 138 ribosomal protein genes (RPGs) [1], which accounts for nearly 50% of all RNA polymerase II (RNAPII) transcriptional initiations [2,3]. In this yeast, the transcription of RPGs is tightly controlled and coordinated forming a transcriptional module [2,3]. It is assumed that RPGs transcription in all species is coordinately regulated forming a gene regulatory network. Since this transcriptional module is rather small and the aminoacidic sequences of RPs are highly conserved, the understanding of the mechanisms that coordinate and regulates this transcriptional module might be useful to understand other transcriptional networks.
The RPGs transcriptional module in S. cerevisiae has been well studied at the cellular level [2,3]. However, although the coding sequences of RPGs are conserved through the eukaryotic kingdom, the promoter sequences controlling their expression are not conserved in different species. For this reason, it is necessary to compare different genomes and identify the ciselements associated with them in each species. It is also necessary to identify the molecular mechanisms of transcription associated with their expression. Another model organism used to study biological process is the fission yeast Schizosaccharomyces pombe, where an RPG transcriptional module has also been described and details of the molecular mechanisms of RPG gene expression have been described. Recently, mechanistic studies using the RPGs promoters from Drosophila have been also performed at some level. In this review, we will summarize and describe the most recent advances in RPGs transcriptional regulation in the above-mentioned model organisms.
Literature Review
Budding yeast Saccharomyces cerevisiae ribosomal protein gene transcription
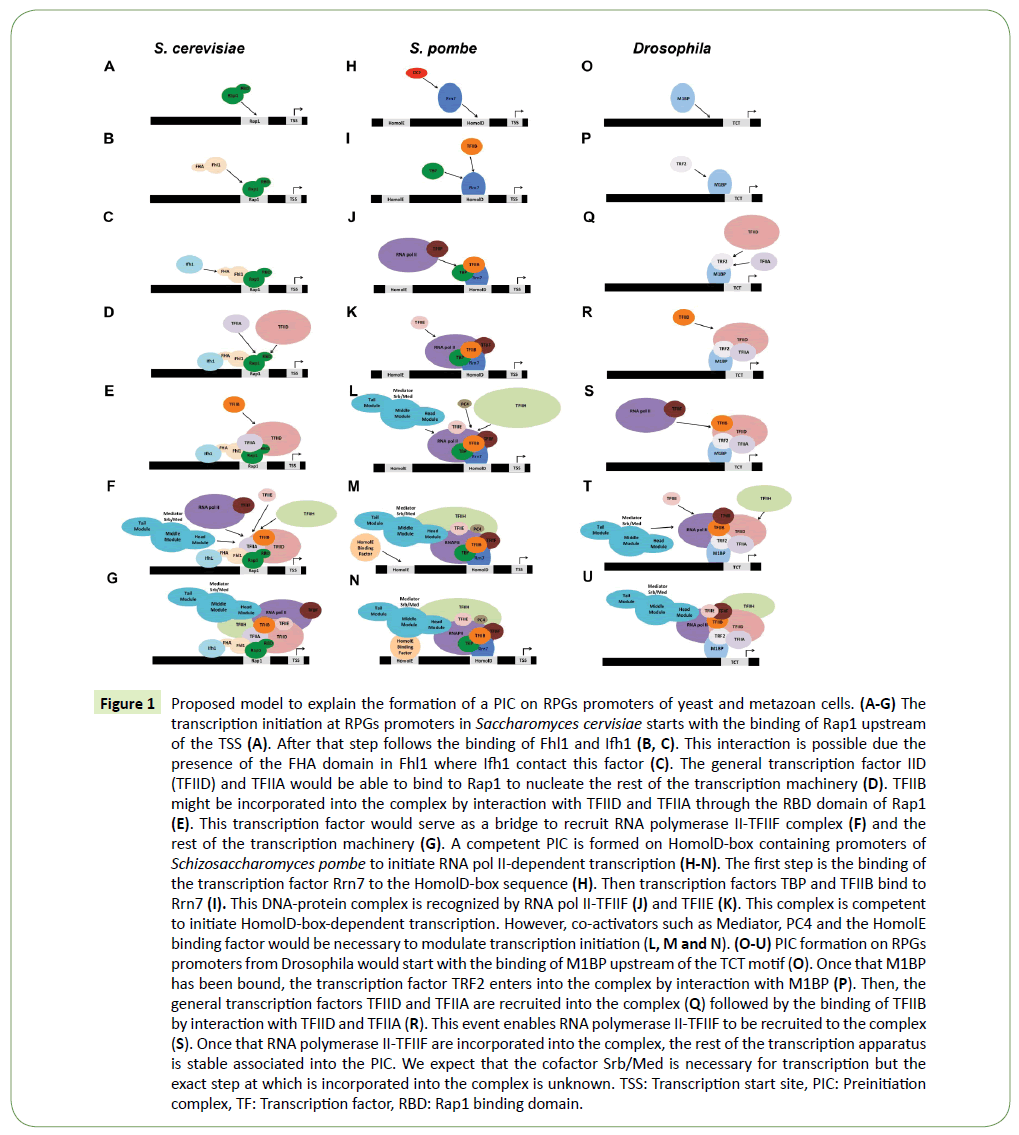
It is known that Saccharomyces cerevisiae contains 79 ribosomal proteins encoded by 138 RPGs which are scattered through the genome, and probably is the most coordinately regulated transcriptional module. The large majority of RPGs promoter contains a cis-element which is able to bind the Repressoractivator- protein 1 (Rap1) [4,5] and it was thought that the coordinated regulation lies on this interaction. However, Rap1 is responsible for the transcription of many other genes including glycolytic and translation factor genes [6,7]. Moreover, Rap1 is required to nucleate complexes to repress transcription of genes adjacent at the telomeres and at the MAT loci [8]. For those reasons, the binding of Rap1 by itself cannot explain the coordinate regulation of RPGs and additional cofactors must specify this specific coregulation. It is known that the cofactors Ifh1 and Fhl1 are critical for RPGs transcription [9,10]. It is currently thought that the first step in RPGs transcription is the binding of Rap1 to its DNA element and clear nucleosomes from the promoter [11]. A second step is the recruitment of Fhl1 (forkhead-like) which in turns is able to recruit Ifh1 through the forkhead (FH)-associated (FHA) domain of Fhl1 and Ifh1 might be able to bind an IFHL DNA motif in some RPGs promoters [9,10]. Thus, the interaction Fhl1-Ifh1 is the responsible to activate RPGs transcription. However, these observations do not explain how the rest of the RNAPII transcriptional machinery is recruited to the RPGs promoters to form a pre-initiation complex (PIC) able to transcribe the gene. Studies from P. Anthony Weil and coworkers [12,13] have demonstrated that Rap1 directly interacts with the general transcription factor TFIID, specifically with Taf4 and Taf5 subunits through the Rap1 binding domains (RBD) which are fundamental for RPGs transcription. Moreover, recently it has been demonstrated that the general transcription factor TFIIA makes essential contributions to the transcription of those genes through contacts with TFIID in the RBD of Taf4 with the N-terminal domain of the Toa2 subunit of TFIIA [14]. Those observations suggest that the binding of Rap1 to the RPGs promoters provides a docking site for the binding of TFIID and TFIIA and that event might serve to recruit RNAPII and the rest of the transcription machinery. That model is outlined in Figures 1A-1G.
Figure 1: Proposed model to explain the formation of a PIC on RPGs promoters of yeast and metazoan cells. (A-G) The transcription initiation at RPGs promoters in Saccharomyces cervisiae starts with the binding of Rap1 upstream of the TSS (A). After that step follows the binding of Fhl1 and Ifh1 (B, C). This interaction is possible due the presence of the FHA domain in Fhl1 where Ifh1 contact this factor (C). The general transcription factor IID (TFIID) and TFIIA would be able to bind to Rap1 to nucleate the rest of the transcription machinery (D). TFIIB might be incorporated into the complex by interaction with TFIID and TFIIA through the RBD domain of Rap1 (E). This transcription factor would serve as a bridge to recruit RNA polymerase II-TFIIF complex (F) and the rest of the transcription machinery (G). A competent PIC is formed on HomolD-box containing promoters of Schizosaccharomyces pombe to initiate RNA pol II-dependent transcription (H-N). The first step is the binding of the transcription factor Rrn7 to the HomolD-box sequence (H). Then transcription factors TBP and TFIIB bind to Rrn7 (I). This DNA-protein complex is recognized by RNA pol II-TFIIF (J) and TFIIE (K). This complex is competent to initiate HomolD-box-dependent transcription. However, co-activators such as Mediator, PC4 and the HomolE binding factor would be necessary to modulate transcription initiation (L, M and N). (O-U) PIC formation on RPGs promoters from Drosophila would start with the binding of M1BP upstream of the TCT motif (O). Once that M1BP has been bound, the transcription factor TRF2 enters into the complex by interaction with M1BP (P). Then, the general transcription factors TFIID and TFIIA are recruited into the complex (Q) followed by the binding of TFIIB by interaction with TFIID and TFIIA (R). This event enables RNA polymerase II-TFIIF to be recruited to the complex (S). Once that RNA polymerase II-TFIIF are incorporated into the complex, the rest of the transcription apparatus is stable associated into the PIC. We expect that the cofactor Srb/Med is necessary for transcription but the exact step at which is incorporated into the complex is unknown. TSS: Transcription start site, PIC: Preinitiation complex, TF: Transcription factor, RBD: Rap1 binding domain.
Transcription of the RPGs in the fission Yeast Schizosaccharomyces pombe
The initial characterization of the promoter sequences of 14 RPGs from the fission yeast Schizosaccharomyces pombe showed discrete conserved modules, which were named Homol A, B, C, D and E [15-17]. These homology regions were completely different from those described in promoters of genes from other yeasts and mammals, such as Rap1 binding sites, TATA-box, Initiator (Inr) or Downstream Promoter Element (DPE). The function of each Homol element was studied out using a promoter-deletion mutants approach [16]. This work showed that the role of Homol A, B, C and E is associated to the function of regulation of transcription initiation, and might have a UAS-like function. Only the HomolD sequence was able to function as an element that could direct transcription initiation in the same way as the TATA-box [15]. The conserved sequence of the HomolD-box is the octamer CAGTCACA/G, however, in several sequences, this element is found in the inverted form as TGTGACTG. The HomolD box is located 39-52 bp upstream transcription start site in the same position as locates the TATA-box in the fission yeast promoters. In an in vivo approach, using reporter-gene assays in S. pombe cells, it was shown that the HomolD-box is necessary to direct and initiate transcription from the RPGs and was postulated to act as a TATA-box analogue and in the same work, using an electrophoretic mobility shift assay (EMSA), a novel protein complex that binds to HomolD-box was identified [15]. In other studies using and in vitro approach, it was shown that point mutations in the HomolD-box sequence abolish completely the ability of this element to direct transcription initiation from the RPGs [18].
Currently, we know that the genome of Schizosaccharomyces pombe contains 141 RPGs encoding the full set of 79 ribosomal proteins. Interestingly, the analysis of the promoter sequences showed that 140 RPGs contained a highly conserved HomolD-box in the region 49 to 104 bp upstream of the ATG start codon [19]. Additionally, other 59 non-RPGs also showed the presence of the HomolD-box in their promoters. In addition, using promoter databases, it was possible to find HomolD-box sequences in several promoters from other eukaryotic organisms, such as humans and plants, indicating the wide distribution of this novel CPE. Interestingly, HomolD-boxes in RPGs promoters are broadly distributed in the Ascomycota fungus phylum [20]. However, in those organisms closely related to the yeast Saccharomyces cerevisiae other CPE, in the same position as the HomolD-box, are present in RPGs promoters. This element is named Rap1 and binds the transcription factor Rap1, which was described previously [21]. It seems likely that Rap1 replaced the HomolDbox of Schizosaccharomyces pombe in Saccharomyces cerevisiae during evolution. Moreover, several other yeast species share both HomolD-box and Rap1 promoter element [20]. The HomolD-box present in the RPGs promoters of the fission yeast is the target of a DNA-binding protein with biochemical features different from TBP. The identification of the HomolD-box binding protein was achieved using DNA affinity chromatography with double-stranded tandem HomolD-boxes covalently attached to a resin. Proteins bound to the resin were eluted and analyzed by mass spectrometry. The result was that the transcription factor Rrn7 was identified in the protein DNA-bound fraction [18]. This factor is a member of the RNAPI transcriptional machinery and its function is to transcribe rDNA in the nucleolus. In the rDNA promoter, this factor is able to bind to a conserved box, which is similar to a HomolD-box. Rrn7 showed a specific HomolD-box binding activity and it is required for the specific transcription of RPGs containing a HomolD-box [18]. Moreover, the GTFs and RNAPII were required for accurate transcription initiation of a HomolD-box containing promoter. Rrn7 is part of the Zn-ribbon protein family related to transcription factor TFIIB, including the mammalian ortholog TAF1B [22]. It possesses a Zn-ribbon domain in the N-terminal region and two cyclin-like domains in the carboxy-terminal region, displaying domain conservation with the TFIIB family members [22]. RPGs which contains a HomolDbox are transcribed by the RNAPII transcription apparatus [18]. The formation of the PIC on a HomolD-box containing promoter was recently described [23]. The first step in the formation of a PIC on these promoters is the binding of Rrn7 to the HomolDbox. As mentioned previously, this step in the PIC establishment might be regulated by phosphorylation of Rrn7 via CK2 protein kinase [24]. Upon binding of Rrn7 to the HomolD-box, the general transcription factors TBP and TFIIB are able to recognize this DNAprotein complex. After the binding of TBP/TFIIB to the complex, the RNAPII/TFIIF complex is recruited, which in turns allows to the TFIIE factor be incorporated into the complex [23]. Finally, Mediator and the coactivator PC4 may be incorporated to the PIC and might modulate basal transcription through a putative HomolE binding factor in those promoters that contain this DNA element. All the steps describing the pathway of complex formation are summarized in Figures 1H-1N.
The expression of genes containing the HomolD-box in their promoters is almost unknown. However, data from analysis of the RPGs expression profiles during several biological processes in S. pombe, e.g. the switch from vegetative to meiotic growth and growth under stress conditions, have revealed a tightly coordinated expression for all 141 RPGs. For example, during the switch from vegetative to meiotic growth, transcription of RPGs is down-regulated, but then within a short time strong reactivation of RPG expression is observed at the beginning of meiosis [19]. The same co-regulation profile is observed in 32 of the 59 non- RPGs that contain a HomolD-box in their core promoter [19]. Many, but not all, of these non-RPG genes encode components whose homologues in other organisms are involved in protein biosynthesis and signal transduction [19].
RPGs from S. pombe have been identified and it is known that individual ribosomal proteins are encoded by two or three related genes whose promoters containing a HomolD-box. Interestingly, in each gene family at least one promoter possesses a tandem repeat ACCCTACCCT or the inverted form (AGGGTAGGGT) upstream of the HomolD-box [16]. This sequence corresponds to the HomolE-box, which is considered a proximal UAS-like sequence for HomolD box-containing promoters, since the presence of this element strongly increases in vivo transcription directed by the HomolD-box [16]. Both promoter elements HomolD and HomolE-boxes must be in the same orientation to be functional. The distance between the boxes is critical in transcription modulation of RPGs and it has been described that the smaller the distance between HomolD and HomolE, the higher the transcription activity. This distance ranges from 0 to 32 nucleotides. Further investigations must be performed to understand how RPGs expression is regulated and which are the mechanisms involved in the coordination between HomolD and HomolE-boxes during RPGs transcription. Although, despite the fact that there are several factors and mechanisms studied in RNAPII-directed transcription, most of the promoters studied possesses a TATA-box, whereas RPGs promoters are TATA-less. Moreover, transcriptional initiation and activation from TATA-less promoters are poorly understood both in metazoan and yeast cells. Thus, the RPGs promoters and the arrangement HomolEHomolD could provide a model to study transcription in TATAless promoters using a promoter element such as HomolD that is analogue to the TATA-box.
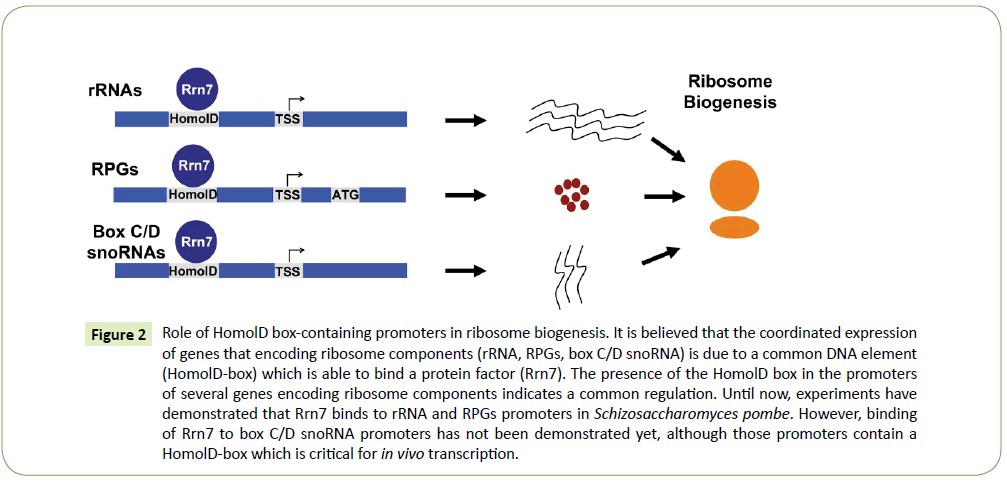
Ribosome biogenesis is one of the most complicated processes in eukaryotic cells, requiring coordinated expression of all ribosome components, which are essential for accurate translation activity. The coordinated regulation and expression of the RPGs with other ribosomal components is still poorly understood. However, in the fission yeast Schizosaccharomyces pombe, it is known that rRNAs, RPGs and box C/D snoRNAs (which are necessary for the modification of rRNAs) contain in their promoters a HomolD-box [18,25], which might be able to control the expression of those genes. Moreover, the Rrn7 transcription factor, which is the HomolD-box binding protein in RPGs, was found to be responsible for the control of the gene expression of box C/D snoRNAs and RPGs in vivo in Schizosaccharomyces pombe cells [25]. Taking all these results together we propose a model, in which the HomolDbox is bound by Rrn7 and co-regulates the transcription of RPG, box C/D snoRNA and rRNA genes in the fission yeast. This model is summarized in Figure 2.
Figure 2:Role of HomolD box-containing promoters in ribosome biogenesis. It is believed that the coordinated expression of genes that encoding ribosome components (rRNA, RPGs, box C/D snoRNA) is due to a common DNA element (HomolD-box) which is able to bind a protein factor (Rrn7). The presence of the HomolD box in the promoters of several genes encoding ribosome components indicates a common regulation. Until now, experiments have demonstrated that Rrn7 binds to rRNA and RPGs promoters in Schizosaccharomyces pombe. However, binding of Rrn7 to box C/D snoRNA promoters has not been demonstrated yet, although those promoters contain a HomolD-box which is critical for in vivo transcription.
Regulation of transcription of the metazoan RPGs
In insects and mammals, RPGs appears to have a common element, which is a member of the poly-pyrimidine initiator (TCT motif). The TCT motif is present in most RPG promoters [26,27] and is required for transcription. The TCT motif encompasses the transcription start site (TSS) and is situated from -2 to +6 bp relative to the TSS, and is therefore located at the same position as the Inr motif, but it cannot function in lieu of an Inr, and it does not bind TFIID. Even so, a single T to A substitution converts the TCT motif into a functionally active Inr [27]. Thus, the TCT motif is a novel transcriptional element which is distinct from the Inr motif. Recently, it has been shown that the TBP-related factor TRF2, but not TBP, is required for the transcription of TCT-dependent RPGs in Drosophila [28]. In Drosophila cells, depletion of TRF2 decreases TCT-dependent transcription. In vitro, recombinant human TRF [29,30] or Drosophila TRF2 are able to activate TCTdependent transcription but not TATA-containing promoters [28]. Nevertheless, TRF2 cannot bind directly to the TCT motif. Thus, another transcription factor must recognize the promoter elements in those RPGs. Indeed, recently a transcription factor named M1BP has been identified as the responsible to bind the core promoter regions of the RPGs and activates transcription of those genes [31]. It has been suggested that M1BP is able to bind to the promoter and recruit TRF2 to the RPGs promoters. Also, the Taf1 subunit of general transcription factor TFIID was present at the RPGs promoters. These observations provide a mechanistic model to envision the formation of a PIC in RPGs promoters of Drosophila. In mammals there is no a counterpart of M1BP, although the TCT motif is present at the RPGs promoters. Whether or not a similar mechanism of RPGs transcription functions in mammals remains to be determined. A model describing the RPGs transcription in metazoans is outlined in Figures 1O-1U.
Discussion and Conclusion
It seems to be clear that the promoter sequences and motifs in the RPGs of different species have evolved; however, the coding sequences have been maintained conserved during the evolution. Perhaps, the transcription of RPGs is controlled by several transcription factors instead of a single one. There are still many unanswered questions about the regulation of RPGs transcriptional regulation. For instance, which are the most frequent cis DNA elements in those genes. How many transcription factors can bind those elements which additional transcription factors or co-activators are necessary to initiate and regulate transcription of RPGs. Is there any crosstalk between the different RNAP machineries that transcribe components of the ribosome.
At least we know that exist a coordinated regulation of transcription in ribosome components of S. pombe where Rrn7 seems to control the transcription of rRNAs, box C/D snoRNAs and RPGs, through the binding of the HomolD box present in the promoters of these genes. The fission yeast Schizosaccharomyces pombe provides an excellent biological model to study the coordinated expression of ribosome components, since the transcriptional regulation of those components seems to be under the control of a few transcription factors. The most immediate questions to answer are:
i) To determine whether or not box C/D snoRNA genes are transcribed by the same transcription apparatus that transcribes RPGs,
ii) To identify the signal that activates transcription of HomolDbox containing genes, and
iii) To identify the HomolE-binding protein. The resolution of all these issues would contribute to understand the regulation of RPGs transcription in the fission yeast and most likely could be extrapolated to metazoan organisms.
Acknowledgment
Work of our laboratory has been fund by the Corporación Nacional de Ciencia y Tecnología de Chile (CONICYT), Fondo de Desarrollo de Ciencia y Tecnología (FONDECYT) and FONDEF (ID16I10145).
References
- Wool IG (1979) The structure and function of eukaryotic ribosomes. Annu Rev Biochem 48: 719-754.
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, et al. (1997) Characterization of the yeast transcriptome. Cell 88: 243-251.
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, et al. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717-728.
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327-334.
- Lascaris RF, Mager WH, Planta RJ (1999) DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics 15: 267-277.
- Morse RH (2000) RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet 16: 51-53.
- Piña B, Fernández-Larrea J, García-Reyero N, Idrissi FZ (2003) The different (sur) faces of Rap1p. Mol Genet Genomics 268: 791-798.
- Moretti P, Freeman K, Coodly L, Shore D (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257-2269.
- Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, et al. (2006) Fine-structure analysis of ribosomal protein gene transcription. Mol Cell Biol 26: 4853-4862.
- Rudra D, Zhao Y, Warner JR (2005) Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24: 533-542.
- Yu L, Morse RH (1999) Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol 19: 5279-5288.
- Layer JH, Miller SG, Weil PA (2010) Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J Biol Chem 285: 15489-15499.
- Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA (2007) Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol 27: 297-311.
- Layer JH, Weil PA (2013) Direct TFIIA-TFIID protein contacts drive budding yeast ribosomal protein gene transcription. J Biol Chem 288: 23273-23294.
- Witt I, Straub N, Käufer NF, Gross T (1993) The CAGTCACA box in the fission yeast Schizosaccharomyces pombe functions like a TATA element and binds a novel factor. EMBO J 12: 1201-1208.
- Witt I, Kwart M, Gross T, Käufer NF (1995) The tandem repeat AGGGTAGGGT is, in the fission yeast, a proximal activation sequence and activates basal transcription mediated by the sequence TGTGACTG. Nucleic Acids Res 23: 4296-4302.
- Gross T, Käufer NF (1998) Cytoplasmic ribosomal protein genes of the fission yeast Schizosaccharomyces pombe display a unique promoter type: A suggestion for nomenclature of cytoplasmic ribosomal proteins in databases. Nucleic Acids Res 26: 3319-3322.
- Rojas DA, Moreira-Ramos S, Zock-Emmenthal S, Urbina F (2011) Rrn7 protein, an RNA polymerase I transcription factor, is required for RNA polymerase II-dependent transcription directed by core promoters with a HomolD box sequence. J Biol Chem 286: 26480-26486.
- Witt I, Kivinen K, Käufer NF (2004) Core Promoters in S. pombe: TATA and HomolD boxes. In: R Egel (ed.). The molecular biology of Schizosaccharomyces pombe. Springer-Verlag Berlin Heidelberg 343-351.
- Tanay A, Regev A, Shamir R (2005) Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA 102: 7203-7208.
- Li B, Nierras CR, Warner JR (1999) Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol 19: 5393-5404.
- Knutson BA, Hahn S (2011) Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors Science 333: 1637-1640.
- Montes M, Moreira-Ramos S, Rojas DA, Urbina F (2017) RNA polymerase II components and Rrn7 form a preinitiation complex on the HomolD box to promote ribosomal protein gene expression in Schizosaccharomyces pombe. FEBS J 284: 615-633.
- Moreira-Ramos S, Rojas DA, Montes M, Urbina F, Miralles VJ, et al. (2015) Casein kinase 2 inhibits HomolD-directed transcription by Rrn7 in Schizosaccharomyces pombe. FEBS J 282: 491-503.
- Diao LT, Xiao ZD, Leng XM, Li B, Li JH, et al. (2014) Conservation and divergence of transcriptional coregulations between box C/D snoRNA and ribosomal protein genes in Ascomycota. RNA 20: 1376-1385.
- Perry RP (2005) The architecture of mammalian ribosomal protein promoters. BMC Evol Biol 5: 15.
- Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, et al. (2010) The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev 24: 2013-2018.
- Wang YL, Duttke SH, Chen K, Johnston J, Kassavetis GA, et al. (2014) TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev 28: 1550-1555.
- Maldonado E (1999) Transcriptional functions of a new mammalian TATA-binding protein-related factor. J Biol Chem 274: 12963-12966.
- Moore PA, Ozer J, Salunek M, Jan G, Zerby D, et al. (1999) A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol Cell Biol 19: 7610-7620.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences