Involvement of a Specific Ubiquitin Ligase in Assembly of the Cytoplasmic Dynein Motor in Neurospora crassa
1Department of Biology, Rockhurst University, Kansas City, USA
2Stowers Institute for Medical Research, Kansas City, USA
3University of Missouri, Kansas City, USA
- *Corresponding Author:
- Ryan Elsenpeter
Department of Biology
Rockhurst University, USA
Tel: 8165014475
E-mail: ryan.elsenpeter@rockhurst.edu
Received date: June 26, 2018; Accepted date: August 06, 2018; Published date: August 13, 2018
Citation: Elsenpeter R, Schnittker R, Plamann M (2018) Involvement of a Specific Ubiquitin Ligase in Assembly of the Cytoplasmic dynein Motor in Neurospora crassa. Genet Mol Biol Res Vol No: 2 Iss No: 2:7
Copyright: © 2018 Elsenpeter R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Eukaryotic cells utilize multiple molecular motor proteins to accomplish intracellular transport. Although there exists numerous forms of kinesin for anterograde transport, only a single cytoplasmic form of dynein carries out the functions of retrograde transport. To accomplish its tasks, dynein makes use of multiple subunits and accessory proteins, including heavy chains, light chains, intermediate chains, light intermediate chains, and dynactin to form a motor complex of several megadaltons.
Methods and Findings: Numerous dynein heavy chain mutants were isolated previously from a genetic screen in the filamentous fungus Neurospora crassa, with a subset located to the C-terminal region, which were the focal point of this work. To explore the mechanism by which these mutations affect dynein function, both intragenic and extragenic suppressors were identified. A novel extragenic suppressor of dynein mutations was discovered, a gene encoding a putative E3 ubiquitin ligase with homologs present in higher organisms, including humans. Mutation or deletion of the suppressor gene results in restoration of wild type-like growth and in vivo dynein localization for each of the C-terminal dynein heavy chain mutants.
Conclusions: Results suggest that mutation of the C-terminal domain of the heavy chain impacts its interaction with dynein intermediate chain. Under this mutated state of dynein heavy chain, a fully functional ubiquitin ligase may act to interfere with proper assembly of the dynein motor.
Background: Eukaryotic cells utilize multiple molecular motor proteins to accomplish intracellular transport. Although there exists numerous forms of kinesin for anterograde transport, only a single cytoplasmic form of dynein carries out the functions of retrograde transport. To accomplish its tasks, dynein makes use of multiple subunits and accessory proteins, including heavy chains, light chains, intermediate chains, light intermediate chains, and dynactin to form a motor complex of several megadaltons.
Methods and Findings: Numerous dynein heavy chain mutants were isolated previously from a genetic screen in the filamentous fungus Neurospora crassa, with a subset located to the C-terminal region, which were the focal point of this work. To explore the mechanism by which these mutations affect dynein function, both intragenic and extragenic suppressors were identified. A novel extragenic suppressor of dynein mutations was discovered, a gene encoding a putative E3 ubiquitin ligase with homologs present in higher organisms, including humans. Mutation or deletion of the suppressor gene results in restoration of wild type-like growth and in vivo dynein localization for each of the C-terminal dynein heavy chain mutants.
Conclusions: Results suggest that mutation of the Cterminal domain of the heavy chain impacts its interaction with dynein intermediate chain. Under this mutated state of dynein heavy chain, a fully functional ubiquitin ligase may act to interfere with proper assembly of the dynein motor.
Keywords
Cytoplasmic dynein; Ubiquitin ligase; Neurospora crassa; Molecular motors; ATPase
Abbreviations
AAA: ATPases associated with diverse cellular activities; DHC: dynein Heavy Chain; DIC: dynein Intermediate Chain; Lis1: Lissencephaly 1; ro-1: ropy-1; SNP: Single Nucleotide Polymorphism; VMM: Vogel’s Minimal Medium; WT: Wild Type
Introduction
To function properly and remain well organized, eukaryotic cells depend on molecular motor systems to transport various cargoes throughout their cellular space. To accomplish their tasks, molecular motors utilize the energy from ATP to travel upon cytoskeletal tracks of either actin filaments or microtubules that not only provide roadways but also overall structure and support for the cell. For more efficient movement of cargoes, different classes of molecular motors will bind to a cargo allowing for both bi-directional movement as well as movement between the actin- and microtubule-based transport systems [1,2].
The bulk of cargo transport occurs along the microtubule lattice by the kinesin and dynein motors. Microtubules have inherent polarity, with plus-ends near the cell periphery and minus-ends at the cell center. Kinesin primarily moves cargoes towards microtubule plus-ends (i.e., anterograde transport), while dynein is involved in retrograde transport, moving cargoes towards the minus-ends of microtubules [3]. dynein, the most complex of the motor proteins, functions as a dimer of heavy chains in concert with various accessory proteins, including dynein intermediate chain (DIC), light chains, the dynactin complex, and lissencephaly 1 (Lis1) [4-6]. The dynein heavy chain (DHC) is one of the largest polypeptides in eukaryotic cells and consists of approximately 4300-4600 amino acids depending upon the organism of origin. DHC is composed of six AAA (ATPases Associated with diverse cellular Activities) domains and contains sites for attachment of the various accessory proteins that assist DHC in accomplishing its tasks. The major accessory complex is dynactin, which is made up of various subunits that aid dynein in both cargo binding and processive movement towards microtubule minus-ends. The other major accessory complex regulating dynein activities is Lis1. Lis1 acts as a regulator of the mechanochemical cycle of dynein and also aids in its microtubule affinity [7-9]. In humans, Lis1 loss-of-function mutations lead to lissencephaly, or smooth brain disorder [10].
This work utilized the filamentous fungus Neurospora crassa to study dynein function. Due to its filamentous growth, this fungus serves as a good model for long-range transport by dynein along microtubules. Many components of the dynein complex are conserved between N. crassa and higher eukaryotes and dynein functions in numerous activities that are common between many species as well [11,12]. dynein mutants display a very distinct and unique growth phenotype, known as ropy, that allows for their easy identification and isolation in the fungus [13,14].
A large-scale screen in N. crassa utilizing a cot-1ts strain was completed to isolate mutants defective in dynein function, including many with mutations in the DHC (ro-1) gene [13,15]. Examination of dynein localization in DHC mutants that produced defective DHC revealed five distinct dynein localization patterns [15]. Amongst these five classes, one class was comprised of six mutants displaying a lightly diffuse, uniform localization pattern in contrast to the wild type (WT) strain, which contains both comet-like structures and a bright, diffuse signal in the rapidly growing hyphal tips [15]. Of these six mutants, four were found to contain mutations that confer changes to amino acids near the C-terminus of the DHC.
With recent structures completed [16-19] some conjecture has been put forth regarding the workings of the C-terminal domain/region. Although numerous studies have been published on the dynein complex, few have been focused on the C-terminal region of DHC. One study found that a DHC motor missing a portion of the protein containing the C-terminal region had a six fold increase in ATPase activity [20], while another created a model whereby the N-terminal tail of DHC slides during the ATPase power stroke, making and breaking contacts between AAA1/2 and the C-terminal domain [21]. The DHC Cterminal domain is highly variable in size between different species, comprised of a small helical region in yeast, with some filamentous ascomycetes adding about 250 residues, while the basidiomycetes and animals have C-terminal domains with about 300 more amino acids than the ascomycetes [22,23]. It now appears that the larger C-terminal domain found in higher organisms, but absent in the ascomycete fungi, works to control both processivity and force production of the dynein motor head [24]. When the C-terminal domain was eliminated from the structure, rat dynein increased its distance traveled along microtubules as well as overall force generation, both comparable to yeast dynein which lacks a C-terminal domain [24]. Interestingly, the C-terminal domain in fungi, including N. crassa, is much shorter than in other higher organisms. This may indicate a different role in overall dynein function in these organisms.
In this study, we report the results of a reversion analysis screen for suppressors of N. crassa dynein mutants. We found that both intragenic lesions as well as extragenic mutations could suppress the C-terminal DHC mutant growth. Thus far, these have been the only DHC mutants that when subjected to reversion analysis contains extragenic suppressor mutations [15]. Examination of extragenic suppressors of dynein C-terminal mutations discovered a gene encoding a putative E3 ubiquitin ligase with homologs present in higher organisms, including humans [25,26]. Mutation or deletion of this suppressor gene results in restoration of wild type-like growth and wild-type in vivo dynein localization for each of the C-terminal dynein heavy chain mutants, as well as certain other dynein heavy chain mutants. Results suggest that the activity of this putative E3 ubiquitin ligase affects interaction of dynein heavy chain with dynein intermediate chain and may be a regulator of overall dynein motor assembly in N. crassa.
Materials and Methods
Culture conditions
Strains were maintained on Vogel’s minimal medium (VMM) with 1.5% sucrose. All handling was completed according to standard technique [27].
Isolation and characterization of DHC mutant strains
Previous studies determined that mutations that reduce or eliminate dynein/dynactin/Lis1 function partially suppress the cot-1ts growth phenotype at 37°C [13,14]. DHC mutant strains were isolated using techniques as previously described [13]. Complementation assays were performed to identify the respective dynein/dynactin/Lis1 gene that was mutated leading to suppression of the cot-1ts growth phenotype. Heterokaryons were created between unknown ropy mutants from the screen and strains with known deficiencies in dynein genes and plated at 25°C. Complementation was defined by the presence of straight hyphal growth for a pair of strains. Noncomplementation was indicated by the presence of ropy growth for the pair upon plating.
In vivo dynein localization studies utilizing dynein fluorescent fusions
Recombinant dynein intermediate chain (DIC) was used for in vivo dynein localization constructed in a separate study [15]. For imaging, standard glass microscope slides were covered with sucrose minimal agar by dipping them in a tube of liquid media. Slides were placed in a Petri dish over two toothpicks for drying. Slides were then inoculated in the center with a small loop of conidia and placed at 28°C overnight (~16-18 hours). Slides were imaged using the Olympus BX50 stereomicroscope (Center Valley, PA) at an exposure of 800, binning of 3 x 3, and a gain of 4. Images were taken using a SPOT RT SE digital camera (Sterling Heights, MI) mounted to the microscope.
UV mutagenesis screen for suppressors of DHC mutants
To identify mutations that suppress the effects of specific DHC mutations, ro-1 mutants were exposed to UV light, plated in minimal agar medium, incubated overnight and survivors examined for revertants displaying wild-type or near wild-type hyphal growth. In brief, conidial suspensions were made in 25 ml sterile water to a count of ~4-5 x 106 conidia/ml. These suspensions were poured into Petri dishes, placed on a table top tray shaker at 150 RPM, and exposed to UV light for 30 seconds which was determined to result in 80%-90% killing. Next, 1l samples of the suspensions were placed into a standard 50 ml screw cap tube and mixed with 30 ml of warm (~50°C) agar. This mixture was then poured into a standard Petri dish and placed at 25°C overnight. Plates were observed under an Olympus SZ-11 Stereo zoom Microscope (Center Valley, PA) and any colonies showing wild-type growth (i.e., those with suppressor mutations) were picked to sucrose minimal slants and incubated at 25°C. These revertants were then streaked out for purity and backcrossed to wild type to eliminate extraneous mutations resulting from UV light exposure.
Identification of intragenic and extragenic suppressors
Revertants deemed pure after UV mutagenesis screening and purification streaking were crossed with wild type to determine the general location of the suppressor mutation(s). Ascospores were gathered after ~10-14 days in 1 ml of water and heatshocked by incubating at 60°C for 30 minutes. The ascospores were then plated on sucrose minimal media plates, incubated at 25°C overnight, and then observed under an Olympus SZ-11 Stereo zoom Microscope (Center Valley, PA) the following day. Strains were separated into sets of intragenic or extragenic suppressor mutation(s) following the screen by backcrossing with wild type. The entire ro-1 structural gene was DNA sequenced for those revertants containing intragenic suppressor mutations.
The Neurospora genome group at Dartmouth developed a novel approach to the discovery of a mutation of interest in the N. crassa genome [28]. The screen was based on tracking single nucleotide polymorphisms (SNPs) that exist between two different laboratory strains of N. crassa, N. crassa Oakridge, the strain used for experimentation throughout this work containing the C-terminal DHC mutations, and N. crassa Mauriceville. Isolation of the region containing the extragenic suppressor mutation(s) of C-terminal DHC mutants was completed using these techniques [28]. To further identify the gene(s) containing suppressor mutation(s), gene deletion cassettes were transformed into C-terminal DHC mutant strains as previously developed and described [27,29]. Any deletion providing a return to wild type growth of transformed strains was sequenced to identify mutations.
Sequencing was processed on an ABI Prism 3100 Genetic Analyzer (Carlsbad, CA) using the manufacturer’s suggested protocol. Sequence reads were analyzed using both the Sequence Scape (ABI, Carlsbad, CA) and Sequencing Analysis (ABI, Carlsbad, CA) programs to check for the presence of mutation(s).
Western blotting of DICm cherry protein
DHC C-terminal mutant and revertant strains were assayed for the presence and level of DICm Cherry protein via m Cherry antibody (1:1000) following the manufacturer’s standard protocol (Novus Biologicals, Littleton, CO).
Results
dynein heavy chain mutant reversion analysis results
In an attempt to gain a better understanding of the function and role of the C-terminal region of DHC, a reversion analysis experiment was completed with four C-terminal DHC mutants (Appendix 1). After completion of UV mutagenesis screening of each of the DHC C-terminal region mutants, strains displaying a return to more WT growth were isolated and purified from any contaminants. Backcrosses with WT strain were completed to determine if the suppressor mutation(s) that arose from exposure to UV light were present in the ro-1 gene or another gene. Genomic DNA from those strains containing intragenic suppressor mutations was prepared and DNA sequencing of the ro-1 gene was completed to identify the mutation(s) present. DNA sequencing resulted in the identification of seventeen unique suppressor mutations of C-terminal DHC mutants (Table 1).
| DHC C-terminal Mutant | Suppressor Mutations | Location in DHC |
|---|---|---|
| G4146A | K1897E | N-terminal tail |
| I2047M | AAA1 | |
| E2278K | AAA2 | |
| Y3905S | AAA5-6 linker | |
| L3916P | AAA5-6 linker | |
| Δ TE3962-3 | AAA5-6 linker | |
| L3985P | AAA5-6 linker | |
| K3992E | AAA5-6 linker | |
| S4364F | C-terminal domain | |
| I4232N | FS 4356 | C-terminal domain |
| D4296E; Δ 4297-99 | V3826I | AAA5-6 linker |
| L3912S | AAA5-6 linker | |
| V4336A | C-terminal domain | |
| FS 4357 | C-terminal domain | |
| P4316S | E1679A | N-terminal tail |
| S4364F | C-terminal domain | |
| A4366T | C-terminal domain |
Table 1: Intragenic suppressors of C-terminal DHC mutants.
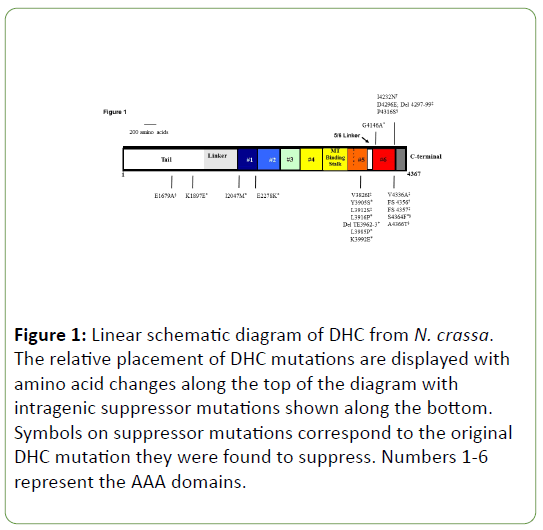
The majority of the intragenic suppressor mutations are found near to the original C-terminal DHC mutations in the five-six linker region (seven) and C-terminal domain (six). Some of these suppressors are able to independently revert each of the four Cterminal DHC mutants and the S4364F suppressor mutation was found for both G4146A and P4316S mutant screens (data not shown). Interestingly, the remaining four suppressor mutations have been mapped to the N-terminal tail, AAA1, and AAA2 region. This area has been shown to lie in close proximity to the C-terminal region when the dynein heavy chain is properly folded in its functional state [16-20]. The majority of intragenic suppressor mutations lie near the C-terminal region of DHC close to the original DHC mutations studied (Figure 1).
Figure 1: Linear schematic diagram of DHC from N. crassa. The relative placement of DHC mutations are displayed with amino acid changes along the top of the diagram with intragenic suppressor mutations shown along the bottom. Symbols on suppressor mutations correspond to the original DHC mutation they were found to suppress. Numbers 1-6 represent the AAA domains.
Reversion analysis of the C-terminal region DHC mutants resulted in both intragenic and extragenic suppressor mutations (Appendix 1). To determine if a suppressor was intragenic or extragenic, the revertant strain was crossed with WT and the spores plated. Any extragenic suppressor strains had a 3:1 ratio of WT growth to ropy growth, while intragenic suppressor strains had only WT growth. To date, this class of mutants is the only that has been found to revert via extragenic lesion(s). One of the C-terminal region mutants (G4146A) was used as the first strain for reversion analysis, which resulted in a disproportionate number of extragenic suppressors in comparison to other Cterminal region mutants. When this is taken into consideration, extragenic suppressors account for about 50% of mutations that lead to reversion of the C-terminal DHC mutants. When isolated suppressor mutation strains are crossed with the various Cterminal DHC mutants, suppression of the ropy growth is seen in all cases, suggesting that these extragenic suppressors can act as universal suppressors of the C-terminal DHC mutants, similar to the phenomenon seen with the intragenic suppressors found at the C-terminal end of DHC (data not shown).
dynein heavy chain mutant and revertant phenotypes
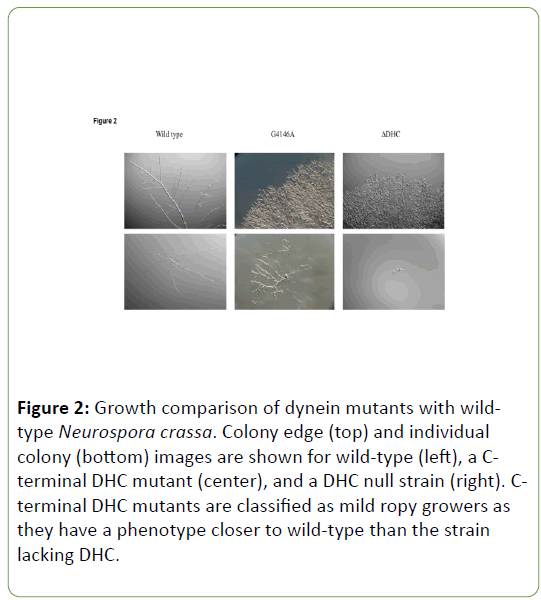
dynein gene mutations display a range of phenotypes, with the strongest being a DHC deletion strain and the weakest being mutants with near WT growth. Those strains with DHC Cterminal region mutations lie in between these two extremes and can be considered to be mild ropy mutants as opposed to more severe ropies like the DHC deletion strain. The difference in growth between WT and C-terminal region mutants is more pronounced when observed for colonial growth than it is for growth from an individual ascospore (Figure 2).
Figure 2: Growth comparison of dynein mutants with wildtype Neurospora crassa. Colony edge (top) and individual colony (bottom) images are shown for wild-type (left), a Cterminal DHC mutant (center), and a DHC null strain (right). Cterminal DHC mutants are classified as mild ropy growers as they have a phenotype closer to wild-type than the strain lacking DHC.
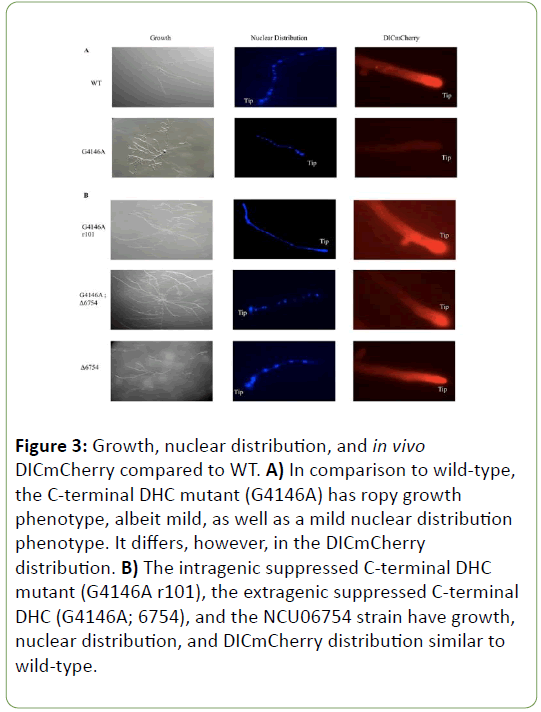
Although C-terminal DHC mutants are considered to have a mild ropy growth phenotype and lack any significant differences for nuclear distribution relative to WT, when examined for their distribution of dynein in the hyphae, they do display a diffuse or haze pattern in contrast to a bright hyphal tip in WT (Figure 3A).
Figure 3: Growth, nuclear distribution, and in vivo DICmCherry compared to WT. A) In comparison to wild-type, the C-terminal DHC mutant (G4146A) has ropy growth phenotype, albeit mild, as well as a mild nuclear distribution phenotype. It differs, however, in the DICmCherry distribution. B) The intragenic suppressed C-terminal DHC mutant (G4146A r101), the extragenic suppressed C-terminal DHC (G4146A; 6754), and the NCU06754 strain have growth, nuclear distribution, and DICmCherry distribution similar to wild-type.
This haze distribution is seen for each of the C-terminal DHC mutants examined for this study. Revertant strains display an almost identical in vivo distribution of motor at the hyphal tip to WT strains as well as a return to more WT growth (Figure 3B). The revertant phenotypes caused by mutation or deletion of the suppressor gene(s) was seen for all DHC strains with AAA6 or Cterminal domain mutations (data not shown).
Single nucleotide polymorphism (SNP) mapping results of extragenic suppressors
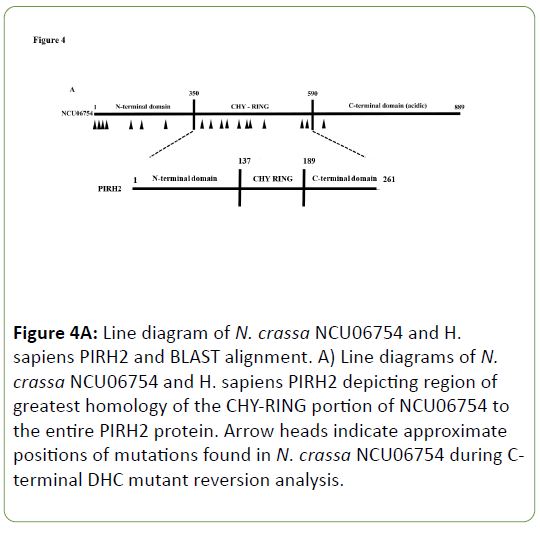
A SNP mapping scheme was developed to locate the extragenic suppressor mutations elsewhere in the genome that when present bring about suppression of the C-terminal DHC mutants ropy growth [28]. By tracing the locations and identities of SNPs in the N. crassa genome, a region on linkage group II was isolated that contains the mutation(s) that lead to suppression of ropy growth for C-terminal DHC mutants. Gene knockouts from this region were then crossed into a C-terminal DHC mutant strain. One, an NCU06754 knockout, brought about suppression of the C-terminal DHC mutant. A total of twenty six revertants showed linkage to this region containing the suppressor and DNA sequencing data was obtained for nineteen different alleles of NCU06754 that act as suppressors of Cterminal DHC mutants (Appendix 2). A number of these alleles contain mutations that confer amino acid changes in and around the CHY-RING of the NCU06754 protein that has a region of homology to the human E3 ubiquitin ligase PirH2 (Figure 4). This region is involved in the coordination of zinc ions via the CHYRING zinc finger and the mutations present in N. crassa NCU06754 map to residues involved in the zinc coordination in human PirH2. Both the N. crassa and human proteins are unique in that they are the only E3 ubiquitin ligases that contain the CHY-RING motif alone without other similar domains [26].
Figure 4A: Line diagram of N. crassa NCU06754 and H. sapiens PIRH2 and BLAST alignment. A) Line diagrams of N. crassa NCU06754 and H. sapiens PIRH2 depicting region of greatest homology of the CHY-RING portion of NCU06754 to the entire PIRH2 protein. Arrow heads indicate approximate positions of mutations found in N. crassa NCU06754 during Cterminal DHC mutant reversion analysis.
Figure 4B: BLAST alignment of region of greatest homology between the two proteins. Arrowheads indicate residues changed in N. crassa that bring about suppression of Cterminal DHC mutants. Conserved residues are indicated by a line between amino acids and residues that have been modified via suppression in N. crassa NCU06754 are shown via arrow heads. Figure 4B created via EMBOSS Water v. 6.6.0 using standard input parameters.
Another minor locus, not linked to this region on linkage group II, has been identified. To date, this locus contains only a single allele. The location of the minor locus was identified via whole genome sequencing. Initial results revealed 556 nonsynonymous SNPs located throughout the genome of the strain, with five ubiquitin-related genes identified amongst the genes containing non-synonymous SNPs. Because many of these gene candidates are essential in N. crassa, the identity of this suppressor was verified by electroporation of a PCR product containing the WT ORF of one of the ubiquitin-related genes, NCU02289, into a C-terminal DHC mutant resulting in a short term RNAi response bringing about a reversion to more WT growth (data not shown). This phenomenon was not seen for the other genes of interest. The suppressor mutation is located within the NCU02289 gene located on linkage group VII, which encodes a putative E2 ubiquitin ligase. Interestingly, the human homolog of NCU02289 has been shown to interact with PirH2, the human homolog of NCU06754 [26].
DICm cherry protein levels do not vary significantly between strains
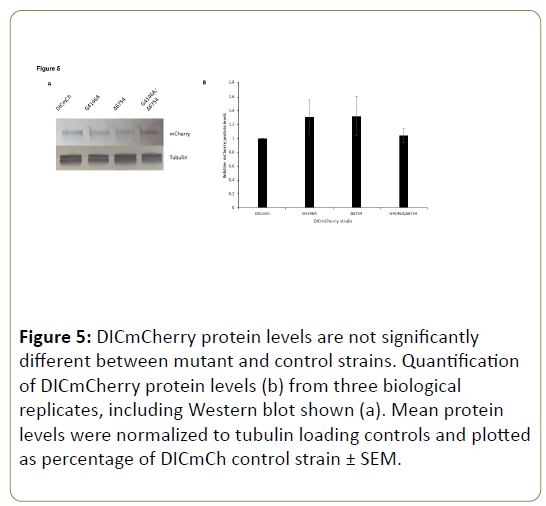
Due to the possibility that DHC C-terminal mutants simply had lower levels of DICmCherry present due to mutation of DHC, Western blots were completed utilizing antibody raised against mCherry protein. When mCherry levels were compared to the DICmCherry control strain, no significant differences were seen in the DHC C-terminal mutant, extragenic suppressor, or suppressed DHC C-terminal mutant strains (Figure 5). Though DICmCherry levels were greatly reduced in vivo when observed in growing hyphae of DHC C-terminal mutants, overall levels of DICmCherry protein were not significantly different from WT when measured via Western blotting.
Discussion
Commonalities of intragenic and extragenic suppressors of c-terminal DHC mutants
Mutants identified from the cot-1 screen were classified on a scale of 1 to 5, with 1 being WT and 5 being a DHC deletion strain. C-terminal DHC mutants are near 3 on the phenotypic scale and are classified as “mild ropies”. The fact that there was little change in nuclear distribution compared to WT further supports these mutants as mild. However, C-terminal mutants have adverse DICmCherry distribution, lacking strong signal near the hyphal tip, rather displaying a diffuse or hazy distribution throughout the hyphae (Figure 3A). This was the first major difference seen compared to WT and seemingly suggested a reduced overall amount of dynein complex present near hyphal tips. Upon completion of reversion analysis, suppressor mutations were identified in the ro-1 gene that brought about a return to more WT growth of C-terminal DHC mutants. In combination, the original C-terminal DHC mutation and an intragenic suppressor mutation bring about a return to more WT growth and an in vivo restoration of dynein localization signal to the hyphal tip (Figure 3B).The majority of these intragenic mutations were found to be located near the site of the original lesion, in and around the C-terminal region (Figure 1, Table 1). There were a few suppressor mutations found elsewhere, notably in the AAA1/2 region of the DHC. This is in agreement with previous work that suggested a relationship between the Cterminal domain and AAA1/2 [20,30]. Because DHC is a ring and conformational changes are propagated throughout the entire ring structure, these suppressor mutations located in other areas may still occur in sites critical for overall dynein structural changes during the ATPase cycle and travel along microtubules. It is also possible that the C-terminal region may be vital for recognition by the ubiquitin ligase system and perturbation of numerous residues and structures in the C-terminus causes DHC to be more preferentially bound by NCU06754, the E3 ubiquitin ligase. By altering this end of the peptide, WT like growth is seen. The relationship between the ubiquitin ligase system and DHC C-terminal suppressor mutations should be a focus of future studies.
Likewise to the intragenic suppressor pattern seen, the extragenic suppressors show commonality in their suppression. Not only does NCU06754 bring about suppression of the Cterminal DHC mutants, but also individual isolated suppressors of C-terminal DHC mutants identified in the NCU06754 gene are able to suppress each of the C-terminal DHC mutants. The same holds for the mutation found in the E2 ubiquitin conjugating enzyme, NCU02289, which was crossed with each of the Cterminal DHC mutants and was able to suppress the ropy growth phenotype for each one.
Model for suppression of DHC C-terminal mutants by modification of NCU06754
Since the major role of E3 ubiquitin ligases is to add ubiquitin groups to proteins thus marking them for proteasomal degradation, an initial model for the suppression of C-terminal DHC mutants by mutation or deletion of NCU06754 is that the original mutation leads to a large increase in degradation of DIC (or another component of the complex) via NCU06754 activity and simply modifying this gene allows for a restoration of protein levels. Based on the in vivo DICmCherry localization data, this hypothesis holds some merit since a hazy, diffuse distribution was seen for in vivo localization (Figure 3A). However, Western blots indicate that DICmCherry protein levels do not significantly vary between strains in this study (Figure 5). This suggests that DIC is present at comparable levels in these strains but when a DHC C-terminal mutation occurs, the motor complex cannot assemble correctly on or near microtubule plus ends near the hyphal tip, leading to a hazy in vivo distribution of DICmCherry.
Figure 5: DICmCherry protein levels are not significantly different between mutant and control strains. Quantification of DICmCherry protein levels (b) from three biological replicates, including Western blot shown (a). Mean protein levels were normalized to tubulin loading controls and plotted as percentage of DICmCh control strain ± SEM.
In this hypothetical model of suppression via mutation or deletion of NCU06754, the E3 ubiquitin ligase targets either an assembly factor for the dynein complex (protein “X”) for degradation or has direct contact with DHC or another dynein subunit or dynein-associated protein (Figure 6). Under normal conditions, DHC is able to properly assemble first with DIC and DLIC because the target protein of the E3 ubiquitin ligase is not polyubiquitinated, which would mark it for degradation. However, under adverse conditions in the cell where there is abnormal dynein structure and/or function due to DHC cterminal mutation, NCU06754 polyubiquitinates its target marking it for degradation and hindering proper assembly of DHC with DIC and DLIC. Another possibility is that under normal conditions, DHC is able to properly assemble first with DIC and DLIC because NCU06754 does not interact with DHC, DIC, or DLIC and does not monoubiqutinate its target which would hinder proper dynein assembly as different states of ubiquitination have been shown to be involved in various biological processes [31]. However, under adverse conditions in the cell where there is abnormal dynein structure and/or function, NCU06754 monoubiqutinates DHC, DIC, or DLIC, which prevents proper dynein assembly. If mutations in the C-terminal region of DHC do not simply reduce the overall level of DIC protein in the cell via NCU06754 and the proteasomal pathway (Figure 5), some other problem must be occurring in the dynein complex brought about by the C-terminal mutations leading to ropy growth and hazy in vivo localization. When the activity of NCU06754 is removed by either mutation or deletion, the mutant dynein complex is seemingly once again able to assemble and localize to the hyphal tips. Future work should be aimed at identifying the cellular target of NCU06754 and the state of ubiquitination as the identity of this protein is critical for better understanding the role of NCU06754 on the dynein complex.
Figure 6: Model for the role of NCU06754 in dynein complex assembly. Dynein complex formation is shown in two steps, first being assembly of DHC with DIC and DLIC then dynein and cargo association along with the addition of Lis1 and dynactin.
Acknowledgement
Strains and plasmids were obtained from the Fungal Genetics Stock Center (Manhattan, KS). The authors would like to thank Dr. Jamie Dyer for discussion and comments on the manuscript.
Funding
This work was supported by grants to M.D.P. (NSF MCB0235871 and NIH P01 GM069087).
References
- Klumpp S, Lipowsky R (2005) Cooperative cargo transport by several molecular motors. Proc Natl Acad Sci USA 102: 17284-17289.
- Ali MY, Lu H, Bookwalter CS, Warshaw DM, Trybus KM (2008) Myosin V and Kinesin act as tethers to enhance each others' processivity. Proc Natl Acad Sci USA 105: 4691-4696.
- Paschal BM, Vallee, RB (1987) Retrograde transport by the microtubule-associated protein MAP 1C. Nature 330: 181-183.
- Hall J, Song Y, Karplus PA, Barbar E (2010) The crystal structure of dynein intermediate chain-light chain roadblock complex gives new insights into dynein assembly. J Biol Chem 285: 22566-22575.
- Hook P (2010) The mechanical components of the dynein motor. Scientific World Journal 10: 857-864.
- Allan VJ (2011) Cytoplasmic dynei. Biochem Soc Trans 39: 1169-1178.
- Yoshida H, Kitagishi Y, Okumura N, Murakami M, Nishimura Y, et al. (2011) How do you RUN on? FEBS Lett 585: 1707-1710.
- Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, et al. (2014) List1 regulates dynein by sterically blocking its mechanochemical cycle. Elife 3: e03372.
- Callejas-Negrete OA, Plamann M, Schnittker R, Bartnicki-García S, Roberson RW, et al. (2015) Two microtubule-plus-end binding proteins LIS1-1 and LIS1-2, homologues of human LIS1 in Neurospora crassa. Fungal Genet Biol 82: 213-227.
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, et al. (1993) Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364: 717-721.
- Seiler S, Plamann M, Schliwa M (1999) Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr Biol 9: 779-785.
- Uchida M, Mourino-Perez RR, Freitag M, Bartnicki-García S, Roberson RW (2008) Microtubule dynamics and the role of molecular motors in Neurospora crassa. Fungal Genet Biol 45: 683-692.
- Plamann M, Minke PF, Tinsley JH, Bruno KS (1994) Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol 127: 139-149.
- Bruno KS, Tinsley JH, Minke PF, Plamann M (1996) Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc Natl Acad Sci USA 93: 4775-4780.
- Sivagurunathan S, Schnittker RR, Razafsky DS, Nandini S, Plamann MD, et al. (2012) Analyses of dynein heavy chain mutations reveal complex interactions between dynein motor domains and cellular dynein functions. Genetics 191: 1157-1179.
- Carter AP, Cho C, Jin L, Vale RD (2011) Crystal structure of the dynein motor domain. Science 331: 1159-1165.
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, et al. (2012) The 2.8 A crystal structure of the dynein motor domain. Nature 484: 345-350.
- Nishikawa Y, Inatomi M, Iwasaki H, Kurisu G (2016) Structural Change in the dynein Stalk Region Associated with Two Different Affinities for the Microtubule. J Mol Biol 428: 1886-1896.
- Schmidt H, Zalyte R, Urnavicius L, Carter AP (2015) Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518: 435-438.
- Hook P, Mikami A, Shafer B, Chait BT, Rosenfeld SS, et al. (2005) Long range allosteric control of cytoplasmic dynein ATPase activity by the stalk and C-terminal domains. J Biol Chem 280: 33045-33054.
- Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M (2007) Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci USA 104: 20832-20837.
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, et al. (2009) AAA+ Ring and linker swing mechanism in the dynein motor. Cell 136: 485-495.
- Numata N, Shima T, Ohkura R, Kon T, Sutoh K (2011) C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett 585: 1185-1190.
- Nicholas MP, Hook P, Brenner S, Wynne CL, Vallee RB, et al. (2015) Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun 6: 6206.
- Corcoran CA, Montalbano J, Sun H, He Q, Huang Y, et al. (2009) Identification and characterization of two novel isoforms of Pirh2 ubiquitin ligase that negatively regulate p53 independent of RING finger domains. J Biol Chem 284: 21955-21970.
- Shloush J, Vlassov JE, Engson I, Duan S, Saridakis V, et al. (2011) Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2. J Biol Chem 286: 4796-4808.
- Davis RH (2000) Neurospora: Contributions of a model organism New York: Oxford University Press 350 p.
- Lambreghts R, Shi M, Belden WJ, Decaprio D, Park D, et al. (2009) A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181: 767-781.
- Collopy PD, Colot HV, Park G, Ringelberg C, Crew CM, et al. (2010) High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol Biol 638: 33-40.
- Imamula K, Kon T, Ohkura R, Sutoh K (2007) The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA 104: 16134-16139.
- Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B (2012) Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 64: 136-142.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences