Insights into Genetic Distances of Pathogenic Leptospira spp. from Humans, Animals and Environment using Multiple Locus Variable-number Tandem Repeat Analysis (MLVA) Genotypes
Sylvia GL1,2*, Luis S1 and Bibiana B1
1Institute of Pathobiology, National Institute of Agricultural Technology, Buenos Aires, Argentina
2National Research Council of Argentina (CONICET), Buenos Aires, Argentina
- *Corresponding Author:
- Sylvia GL
Institute of Pathobiology
National Institute of Agricultural Technology
Hurlingham, Buenos Aires, Argentina
Tel: 0054-11-46211289
E-mail: grune.sylvia@inta.gob.ar
Received Date: November 11, 2016; Accepted Date: December 14, 2016; Published Date: January 02, 2017
Citation: Sylvia GL, Luis S, Bibiana B. Insights into Genetic Distances of Pathogenic Leptospira spp. from Humans, Animals and Environment using Multiple Locus Variable-number Tandem Repeat Analysis (MLVA) Genotypes. J Zoonotic Dis Public Health. 2017, 1:1.
Abstract
Title: Insights into genetic distances of pathogenic Leptospira spp. from humans, animals and environment using Multiple Locus Variable-number tandem repeat Analysis (MLVA) genotypes.
Background: Leptospirosis is the most widespread zoonosis worldwide. This disease is caused by bacteria, pathogenic strains of the genus Leptospira spp. belonging to the order Spirochaetales. These pathogenic strains of Leptospira spp. are disseminated through urine of infected animals and/or reservoirs into the environment. Human and animal vaccines are not available in many countries and are serovar specific. It is very important to study the genotypes of pathogenic leptosprial strains, since this allows knowing the current epidemiological scenario from endemic countries and in high risk areas.
Methods and findings: A total of 67 pathogenic Leptospira spp. strains were used in this study. Genotyping using Multiple Locus Variable-number tandem repeat Analysis (MLVA) was carried out. The discriminatory power of two sets of loci (Set A: VNTRs 4, 7, 9, 10, 19, 23, 31 and Set B: VNTRs: 4bis, 7bis, 10bis, Lb4, Lb5) was analyzed by Principal Coordinate Analysis (PCoA). This type of analysis has not been previously applied to bacterial genotypes. PCoA was done using a distance matrix of the genotypes obtained and the genetic variability was analyzed. According to the numbers of alleles found, it was possible to conclude that the discriminatory power of the five (5) loci of VNTR Set B is higher than that of the seven (7) loci of VNTR Set A. The discriminatory power of VNTR Lb5 and VNTR 31 is very high, since with one different allele the genotypes characterized in this study could be separated.
Conclusion: In this study the use of PCoA was very helpful to analyse the obtained pathogenic Leptospira spp. genotypes isolated from animals, humans and the environment. This analysis gave a current results compared with recent studies that include the evaluation of the discriminatory power of VNTRs for genotyping of pathogenic leptospiral strains. The VNTR Lb5 and VNTR 31 have the greatest power of discrimination between serovars.
Keywords
Leptospirosis; Pathogenic; MLVA; Genotypes; Animal-Human-Environment interfase
Introduction
Leptospirosis is caused by pathogenic strains of the genus Leptospira spp. This bacterium belongs to the order Spirochaetales and infects humans and animals. Pathogenic strains of Leptospira spp. are disseminated through urine of infected animals and/or reservoirs into the environment. This disease can be occupational or recreational since the ways of infection can be direct or indirect. This zoonosis is the widest ranged disease in the world and is endemic in tropical and subtropical countries [1]. The World Health Organization (WHO 2016) estimates more than 1 million cases of human leptospirosis worldwide each year and 58,900 cases with fatal results. In Argentina, leptospirosis is endemic with epidemic outbreaks in humans and animals. Infections of cattle provoke important economic losses due to abortions, stillbirth’s birth of weak animals, infertility and mastitis with reduction of milk production. It is difficult to estimate the losses caused by this concept because inherent diagnostics of this disease [2]. Rapid diagnostic test are not early leptospirosis diagnostic test. Performance of diagnostic tests are frequently expressed in terms of specificity, sensitivity and predictive values [3], this means that in house techniques and innovations have to developed in laboratories that work under good practice guidelines.
Leptospirosis presents a major challenge both in humans and animals since this disease can be controlled but not eradicated. The severity of this disease varies with the infecting serovar and the affected species. It has been found in almost all areas of the world, with the exception of the Polar Regions [1].
The committee of Taxonomy of Leptospira spp. of the International Union of Microbiological Societies and the subcommittee of taxonomy of the International Leptospirosis Society (ILS) approved the following nomenclature for serovars of Leptospira spp.: the genus and species must be italicized and the serovar name (nor italicized) must have an upper case first letter, for example: Leptospira interrogans serovar Icterohaemorrhagiae or L. interrogans serovar Icterohaemorrhagiae. Leptospires are further classified into serogroups and serovars, which can be represented by different species of leptospires, for example the 14 serovars described for Bataviae are represented by L. interrogans (five (5) serovars), L. santarosai (five (5) serovars), L. kirschneri (one (1) serovar), L. noguchii (two (2) serovars) and L. borgpetersenii (one (1) serovar). This reflects a huge taxonomic problem since the incorporation of molecular typing tools show that serovars are shared by different species and no serotyping can represent this.
At the last meeting of the ILS (2015) in Semarang (Indonesia), the subcommittee announced that the definition of new species is based now on in silico hybridization of whole genome sequences, and that the serovar definition is still not possible using only genome sequences [4]. In this meeting, the species L. mayottensis (isolated from humans in the Indian Ocean) was recognized as a new pathogenic species [5]. In a recent study of several Leptospira spp. genomes [6], a comparison between different genes (housekeeping, highly conserved and highly variable) used as typing tools, revealed the grouping into the three known clusters. The tree constructed based on secY, the universal markers and the pan-genome clearly separated the three pathogenic species L. interrogans, L. kirschneri and L. noguchii from the other six pathogenic species. Interestingly, the tree constructed on 16S rRNA did not discriminate between L. meyeri and L. yanagawae, being L. meyeri in some studies considered as a pathogenic species and in other studies as saprophyte, this seems to depend on the current gene selection Correlation between genetic diversity and biological features is still pending. The relationship between the genomes of Leptospira spp., the pathogenicity and the ability to survive in environmental niches is an especially important area of investigation.
The major infections of global importance are serovar Icterohaemorrhagiae infection in rats, serovar Hardjo in cattle and sheep and serovar Canicola, and possibly serovar Bratislava, in pigs and dogs. Also infection of pigs by serovars Kenniwicki or Tarassovi have more limited geographical spread either due to limitations in host distribution or to unrecognized factors [2].
Several molecular techniques have been used to characterize isolated strains from clinical samples, including MLVA, MLST, and sequencing of the 16S rRNA and sec Y genes.
The purpose of this study is to analyze the genetic distance by Principal Coordinate Analysis (PCoA) of MLVA characterized genotypes of pathogenic Leptospira spp. strains isolated from animals (domestic and wildlife), humans and the environment (river) and to evaluate the discrimatonry power of the VNTRs used in Set A and Set B in this study. The objective is to use fewer VNTR to genotype pathogenic strains.
Materials and Methods
Culture of strains
A total of 67 strains were analyzed in this study. The sources date and locations of isolation of each strain are listed in Tables 1 and 2. Thirty-one (31) of these were recent isolates (2010-2014) and 36 were part of the bacterial collection of the Leptospirosis Laboratory at the Pathobiology Institute of the Argentine National Institute of Agricultural Technology (1961-2009), which is an OIE reference laboratory. The isolates used in this study were previously serotyped using the Cross- Agglutinin Absorption Test (CAAT). The reference Leptospira interrogans strains employed in this study were Pomona (serovar Pomona, serogroup Pomona), M20 (serovar Copenhageni, serogroup Icterohaemorrhagiae), RGA and Ictero No. 1 (serovar Icterohaemorrhagiae, serogroup Icterohaemorrhagiae), Hond Utrecht IV (serovar Canicola, serogroup Canicola) and MY 1039 (serovar Portlandvere, serogroup Canicola). The reference strain used for Leptospira borgpetersenii was Castellon 3 (serovar Castellonis, serogroup Ballum). Reference strains and isolates were grown in Fletcher medium (Difco Laboratories) at 28°C as described before [7].
| Serovar | MLVA genotype (results for VNTR4, VNTR7, VNTR9, VNTR10, VNTR19, VNTR23 and VNTR31 loci) | Number of isolates/source | Reference |
|---|---|---|---|
| Icterohaemorrhagiae | "Ictero" (2,1,13,7,2,0,3) | 5/rats | [7] |
| 2/bovine | [14] | ||
| 1/river | [12] | ||
| 3/humans | [12] | ||
| ”Ictero-like” (2,1,10,7,2,0,3) |
1/fox | [16] | |
| Subtotal | 12 | - | |
| Pomona | Pomona (2,0,6,14,8,1,3) | 1/bovine | [10] |
| Pomona genotype A (4,1,6,10,8,2,3) | |||
| 1/human | [10] | ||
| Pomona genotype C (6,1,6,10,8,2,3) | 1/human | [10] | |
| “Pomona-like” (0,6,14,8,1,3) | 1/canine | [13] | |
| Subtotal | 4 | - | |
| Canicola | Hond Utrecht IV (1,10, 2, 3, 10, 2,3) |
1/squirell | [14] |
| 1/skunk | |||
| 3/opossum | [14] | ||
| 2/bovine | [14,12] | ||
| 2/rats | [7] | ||
| 2/humans | [12] | ||
| 2/canines | [13] | ||
| Subtotal | 13 | ||
| Portlandvere | MY 1039 (1,10, 2, 3, 10, 2,4) | 1/rat | [13] |
| 2/bovines | [12] | ||
| 4/canines | [7] | ||
| Subtotal | 7 | - | |
| Muelleri | RM2 (2,3,6,7,1,6,-) | - | - |
| RM2-like (2,3,6,7,1,6,3) | 1/rat | [14] | |
| Subtotal: | 1 | - | |
| Total | 37 | - |
Table 1: Multiple locus variable-number tandem repeat analysis (MLVA) genotypes found in different hosts using Set A (7 VNTR loci).
| Serovar | MLVA genotype (results for locus: VNTR4, VNTR7, VNTR10, VNTRLb4, VNTR Lb5) | Isolations/source | Reference |
|---|---|---|---|
| Castellon | Castellon 3 (1,-,1,6,7) | 4/rats | [7,14] |
| 2/mice | [7,14] | ||
| 1/opossum | [14] | ||
| 1/ovine | [14] | ||
| 1/wild boar | [14] | ||
| Subtotal | 9 | - | |
| Icterohaemorrhagiae | Ictero-1 (2,1,7,0,4) | 6/rats | [7,14] |
| 1/squirell | - | ||
| RGA (2,1,7,0,0) | 1/canine | [13] | |
| Subtotal | 8 | - | |
| Copenhageni | Fiocruz L1-130 (2,1,7,0,6) | 10/rats | [14] |
| 1/mouse | [14] | ||
| 2/opossum | [14] | ||
| Subtotal | 13 | - | |
| Total of isolates (5 loci) | 30 | - |
Table 2: Multiple Locus Variable-number tandem repeat Analysis (MLVA) genotypes found in different hosts using Set B (5 VNTR loci).
Genotyping
Multiple Locus Variable-number tandem repeat Analysis (MLVA) typing was performed using two sets of oligonucleotides specific for pathogenic L. interrogans, L. kirschneri and L. borgpetersenii. Oligonucleotides that hybridized to the flanking regions of the VNTRs: 4, 7, 9, 10, 19, 23 and 31 loci (Set A) were used to discriminate strains belonging only to L. interrogans [8] and oligonucleotides that hybridized to the flanking regions of the VNTRs: 4bis, 7bis, 10bis, Lb4 and Lb5 loci (Set B) were used for L. kirschneri, L. borgpetersenii and L. interrogans strains [9]. The MLVA strain typing procedure was performed as described [8-14]. DNA was extracted from 20 μl of culture using Chelex-100 resin (Bio Rad) [14]; extracted DNA was stored at -20°C until use.
The final volume (50 μL) of each reaction mixture contained polymerase chain reaction (PCR) buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCL), 200 μM deoxynucleoside triphosphates, 2 μM each of the corresponding forward and reverse primers, 2 mM MgCl2, 1.25 U of Taq DNA polymerase (Invitrogen) and 5 μL of DNA template. PCR amplifications were carried out in a Thermo Scientific PxE 0.2 Thermal Cycler, using the following cycling parameters: 94°C for 5 min, followed by 35 cycles of denaturalization at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 90 s, with a final cycle at 72°C for 10 min. The amplified samples were examined following electrophoresis in ethidium bromide-containing 2% agarose gel in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) at 100 V for 50 min. Amplified DNA bands were visualized through ultraviolet light exposure (Uvi Tec transiluminator BTS-20.M, Manufacturer UviTec, St. John’s Innovation Centre, Cowley Road, Cambride, England). Amplicon sizes were estimated using CienMarker (Embiotech, Buenos Aires, Argentina) and the Gel Analyzer 2010a program. To calculate repeat copy numbers, the following formula was used: number of repeats (bp)=[fragment size (bp)-flanking regions (bp)]/ repeat size (bp). Repeat copy numbers were rounded down to the closest whole number. If the copy number was less than one, it was rounded to zero [10].
Analysis of genetic distances
Once the genotypes were characterized, genetic distances between the isolates were determined. The Excel complement GenAlEx was used to calculate a triangular distance matrix as a previous step to the Principal Coordinate Analysis (PCoA). In the PCoA a centered matrix is calculated and then decomposed into its component eigenvalues and eigenvectors. The eigenvectors standardized by dividing by the square root of their corresponding eigenvalue, are output as the principal coordinate axes. This analysis is also called metric multidimensional scaling. It is useful for ordination of multivariate data on the basis of any distance function [15]. In this study we used the genetic distances of pathogenic Leptospira spp. genotypes. Gower [15] created PCoA this analysis is used mainly in statistics. Two PCoA were carried out, one for seven (7) loci (only for genotypes belonging to L. interrogans) (Figure 1) and the other for five (5) loci (used for genotypes belonging to L. interrogans, L. borgpetersenii and L. kirschneri) (Figure 2). The number of alleles is the same as the copy numbers of each flanked VNTR. For example for the locus VNTR4 of a particular MLVA genotype the copy number obtained is two (2), this means that the number of alleles for this VNTR4 is also two (2). This is relevant for the interpretation of genetic distances.
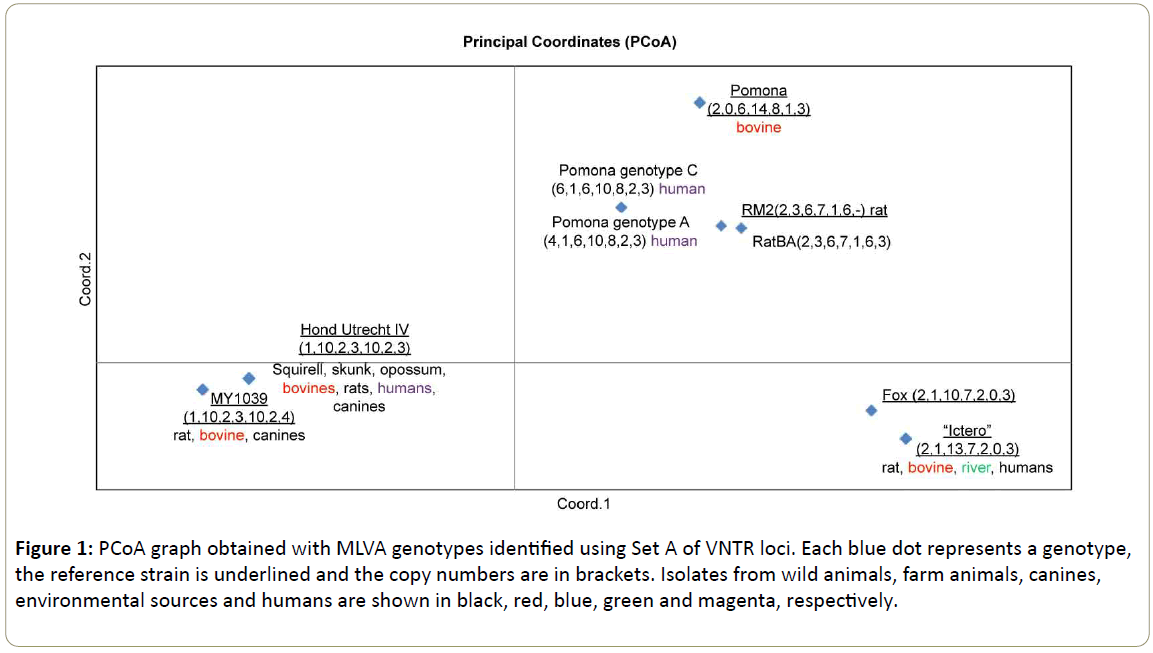
Figure 1: PCoA graph obtained with MLVA genotypes identified using Set A of VNTR loci. Each blue dot represents a genotype, the reference strain is underlined and the copy numbers are in brackets. Isolates from wild animals, farm animals, canines, environmental sources and humans are shown in black, red, blue, green and magenta, respectively.
Results
Tables 1 and 2 shows the MLVA genotypes of each characterized sample using VNTRs of Set A and B, respectively. A total of 37 strains could be characterized using Set A. The best represented genotype was the “Ictero” group with copy numbers (2,1,13,7,2,0,3) belonging to 11 isolates (five (5) wildlife, two (2) bovines, three (3) humans and one (1) river). A similar genotype was determinated from an isolated strain of a fox (South American Gray Fox) represented with the copy numbers (2,1,10,7,2,0,3) [16], the allelic difference (+3) was determinated in the VNTR9. The representation of this group is shown in Figure 1 on the right lower side.
Representing the genotypes of serovar Pomona, only four (4) strains were included in this study, since in a previous study, four genotypes belonging to serovar Pomona were established [10]. Analyzing Pomona genotype C (6,1,6,10,8,2,3) and Pomona genotype A (4,1,6,10,8,2,3) we can observe an allelic difference (+2) in VNTR 4, this loci does not discriminate between genotypes, both genotypes C and genotype A of Pomona are represented by the same blue dot in the PCoA (Figure 1), however the genotype Pomona Pomona represented by copy numbers (2,0,6,14,8,1,3) can be found in the same upper right square of the PCoA but not closely related to genotypes A and C of Pomona.
The genotype of serovar Canicola Hond Utrecht IV (1,10,2,3,10,2,3) was well represented with 13 isolations (two (2) humans, seven (7) wildlife animals and four (4) domestic animals). This genotype is represented in the PCoA (Figure 1) in the lower left square closely related to the genotype of serovar Portlandvere MY 1039 (1, 10, 2, 3, 10, 2, 4), this genotype was found in seven (7) isolated strains (animals). The allelic difference between both serovars Canicola and Portlandvere is (+1) at the VNTR31 loci.
In Grune et al. [13], MLVA genotypes of rodent populations in urban and periurban regions of Buenos Aires were presented. In this study we included a strain with following copy numbers (2,3,6,7,1,6,3), that was similar to the genotype serovar Grippothyphosa RM2 (L. interrogans) with copy numbers (2,3,6,7,1,6,-). In this case we can observe that the allelic difference in VNTR 31 is (+3). Surprisingly the serovar Grippothyphosa (L. interrogans) seems to be closely related to the serovar Pomona, since it is represented in the same upper right square of the PCoA as serovar Pomona (Figure 1).
Summarizing these results, the genotypes belonging to the species L. interrogans were genotyped using Set A (7 VNTR loci) [8]. We found that the VNTR 9, VNTR31 have discriminatory power and VNTR 4 does not. In the PCoA (Figure 1), we could observe the closeness of serovars belonging to the serogroup Canicola and the closeness of serovar Pomona with serovar Grippothyphosa. The more distanced serogroup was Icterohaemorrhagiae.
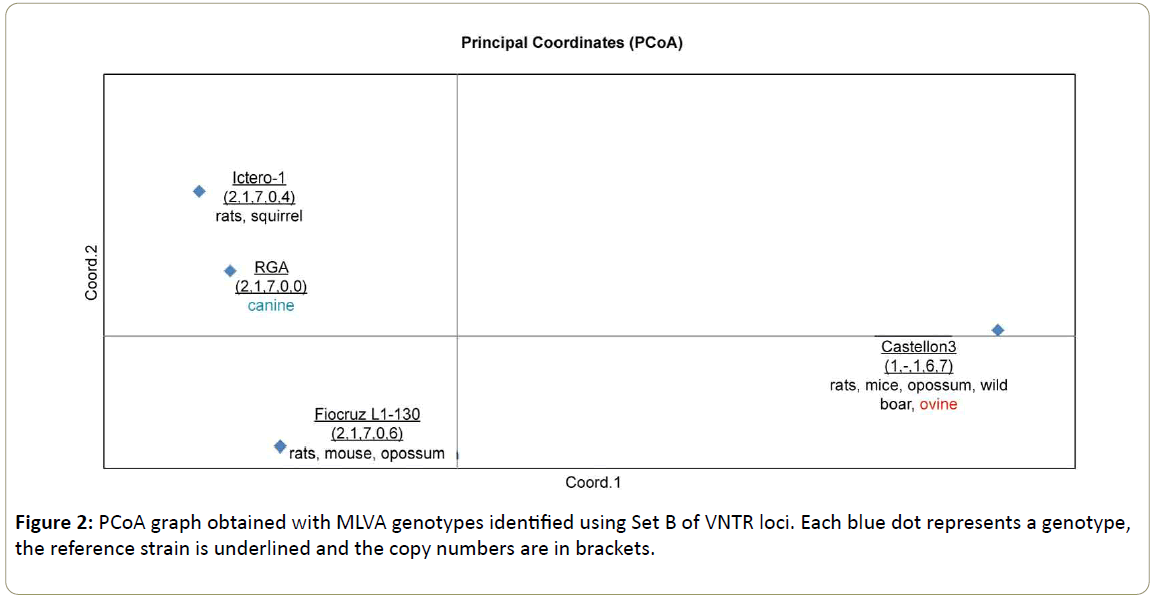
Table 2 lists the genotypes listed obtained using Set B (5 VNTR loci: 4bis, 7bis, 10bis, Lb4, Lb5) [9]. A total of 30 isolates could be characterized with this set. Genotype Castellon 3 with copy numbers (1,-,1,6,7) was represented by nine (9) isolated strains (wildlife and domestic animals). This genotype can be found in the PCoA (Figure 2) in the lower left square, far away from the genotypes belonging to the serogroup Icterohaemorrhagiae represented by serovar Copenhageni Fiocruz L1-130 and serovar Icterohaemorrhagiae RGA and Ictero-1. This reflects that the serovars of the serogroup Icterohaemorrhagiae belong to the species L. interrogans (upper and Lower Square of the PCoA in Figure 2 and the serovar Castellonis Castellon 3 belongs to the species L. borgpetersenii.
Representing the serovar Icterohaemorrhagiae (isolated in animals), we could discriminate two genotypes Ictero-1 with copy numbers (2,1,7,0,4) and RGA with copy numbers (2,1,7,0,0). The allelic difference between both genotypes is (+4) in VNTR Lb5. These genotypes are grouped in the upper left square of the PCoA (Figure 2). With only an allelic difference of (+2) in the same VNTR Lb5, the genotype Fiocruz L1-130 representing the serovar Copenhageni could be discriminated from both genotypes of the serovar Icterohaemorrhagiae placing this genotype in the lower left square, far away from the genotypes of the genotypes Ictero-1 and RGA.
The 12 strains characterized as "Ictero" (2,1,13,7,2,0,3) in Table 1 and Figure 1, could not be genotyped with Set B, since cultures did not grow efficiently. In this case they were classified in the "Ictero" group without discriminating between serovars.
The results obtained reflect the accuracy of carrying out PCoA with genotypes of MLVA-characterized pathogenic Leptospira spp. strains. In this study we could observe that the VNTR loci with the highest discriminatory power are VNTR 9, VNTR31 and VNTRLb5.
Discussion
PCoA is an eigenanalysis algorithm like Principal Component Analysis (PCA), the difference is that PCA only uses the Euclidean distances between objects to perform an ordination in reduced space and PCoA performs an ordination on any user-selected dissimilarity measure. Recently, PCoA has been used to analyze multivariable datasheets based on distance matrixes of genotypes belonging to animal taxa [17] and microbial diversity [18]. This is the first time PCoA is used to analyze bacterial genotypes.
Previous studies describing and comparing molecular typing tools for pathogenic Leptospira spp. describe MLVA as a powerful tool with noteworthy discriminatory capacity [8-21]. In a recent study [19] describe a simple, rapid and robust genotyping High Resolution Melting (HRM) method to discriminate between pathogenic Leptospira spp. species and subspecies. In this work VNTR-Lb5 and VNTR-4bis were used for L. interrogans, VNTR Lb4 and VNTR Lb5, for L. borgpetersenii and VNTR-4bis and VNTR-Lb5 for L. kirschneri. Apparently, VNTR Lb4 can only discriminate L. borgptersenii genotypes [6,19]. In the study at hand, discrimination between L. borgpetersenii genotypes could not be carried out because of lack of isolates [19] could determine that VNTR Lb5 has high discriminatory power using HRM. This result coincides with our observations that showed that VNTR Lb5 was one of the VNTRs with highest discriminatory power using MLVA.
Zilber et al. [22] compared VNTR analysis with Multispaces Sequence Typing (MST) and found that MST could be a new genotyping method for the epidemiological surveillance of L. interrogans; MST could discriminate between serovars of the serogroup Icterohaemorrhagiae.
In this study, using PCoA, we found that, the discriminatory power of VNTR Lb5 and VNTR 31 is very high, since genotypes of different serovars could be differentiated based on one different allele, or example: Serovar Copenhageni from serovar Icterohaemorrhagiae, and serovar Canicola from serovar Portlandvere. Also, but with higher allelic difference (+3) in VNTR9 we could find a similar genotype belonging to the group "Ictero", represented by the “fox” genotype On the other hand, genotypes Pomona A and Pomona C could not be distinguished using PCoA, this reflects that VNTR 4 does not discriminate between genotypes.
It is important to incorporate the analysis of genetic distances when a new or improved molecular typing tool is elaborated and observe the genetic pattern in a pool of isolated pathogenic strains. Using this kind of analysis, the coincidence of genetic profiles from different sources can be visualized in a graphic and the common profiles can be analyzed by region, year of isolation, source, etc. This is particularly important in zoonotic diseases since environmental niches are often used by the bacteria as a travelling method, maintenance and proliferation. In this study the best represented genotype was serovar Canicola Hond Utrecht IV (Table 1), being isolated from 2 humans, 7 wildlife animals and 4 domestic animals. Grune et al. genotyped Leptospira spp. isolates from dogs and reported that the current vaccine used in Argentina does not include all genotypes present in the region [7,13,14]. Until now, and worldwide, this genotype has not been isolated from environmental sources. But, on the other hand, the group of “Icteros” was represented by isolates from humans, animals and the environment (1 strain isolated from a river). Unfortunately, these strains could not be used further for MLVA typing, because of the loss of the cultures.
Koizumi et al. [20] used MLVA genotyping to characterize isolates from wildlife in Japan. In this study, different VNTR loci than those used here were described to genotype serovars from the species L. borgpetersenii (VNTR-Lb4, VNTR Lb-5, V-LbJ19, V-LbL15, V-LbL23). Koizumi et al. [21], indicate that, MLVA has greater genetic divergence than other molecular typing tools. Indeed, their results indicated that MLVA had a higher discriminatory power and was more concordant with serotyping than MLST.
Childeroli et al. [23] characterized L. borgpetersenii serovar Hardjo strain Hadjobovis isolates, differentiating them from Leptospira interrogans serovar Hardjo strain Hardjoprajitno using MLVA and sec Y sequencing. Some authors have indicated that VNTR analysis could lead to ambiguous results in size determination by gel electrophoresis [24] and some serovars could not be discrimiated.
One limitation of MLVA typing is the relatively high amount of genomic DNA needed which restricts its use to isolated strains and hinders its application to clinical samples.
Conclusion
We analyzed the discriminatory power of two (2) sets of VNTR (Set A and Set B) used to genotype pathogenic strains of Leptospira spp. used in MLVA. In our study we analyzed the dissimilarity power using PCoA of VNTRs (Set A) (Set B) [8,9], and found that VNTR31, VNTRLb5 have the highest discriminatory power. The objective is to use fewer VNTR to discriminate all serovars. This reduces costs and helps to create a simple to use molecular typing tool.
Acknowledgment
This research was financed by INTA (AESA 202821). Sylvia Grune has a postdoctoral CONICET scholarship. We are especially grateful to Dr. Leonardo Campagna who contributed the original idea of using PCoA for pathogenic Leptospira spp. strains. We also thank Dr. Monica Jacobsen for the revision of this paper.
References
- Adler B (2015) History of leptospirosis and Leptospira. In: Adler B (ed.) Leptospira and Leptospirosis. Curr Topics Microbiol Immunol 387: 12.
- Ellis W (2015) Animal leptospirosis. In: Adler B (ed.) Leptospira and Leptospirosis. Curr Topics Microbiol Immunol 387: 99-134.
- Picardeau M, Bertherat E, Jancloes M, Skouloudis AN, Durski K, et al. (2014) Rapid tests for diagnosis of leptospirosis: Current tools and emerging technologies. Diagn Microbiol Infect Dis 78: 1-8.
- Levett PN, Picardeau M (2016) International Committee on Systematics of Prokaryotes. Subcommittee on the taxonomy of Leptospiraceae. Int J Syst Evol Microbiol.
- Bourhy P, Collet L, Brisse S, Picardeau M (2014) Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int J Syst Evol Microbiol 64: 4061-4067.
- Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, et al. (2016) What Makes a Bacterial Species Pathogenic?: Comparative Genomic Analysis of the Genus Leptospira. PLoS Negl Trop Dis 10: e0004403.
- Grune Loffler S, Pavan ME, Vanasco B, Samartino L, Suarez O, et al. (2014) Genotypes of pathogenic Leptospira spp. Isolated from rodents in Argentina. Mem Inst Oswaldo Cruz 109: 163-167.
- Majed Z, Bellenger E, Postic D, Pourcel C, Baranton G, et al. (2005) Identification of variable-number tandem-repeat loci in Leptospira interrogans sensu stricto. J Clin Microbiol 43: 539-545.
- Salaün L, Mérien F, Gurianova S, Baranton G, Picardeau M (2006) Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J Clin Microbiol 44: 3954-3962.
- Pavan ME, Cairó F, Brihuega B, Samartino L (2008) Multiple-locus variable-number tandem repeat analysis (MLVA) of Leptospira interrogans serovar Pomona from Argentina reveals four new genotypes. Comp Immunol Microbiol Infect Dis 31: 37-45.
- Pavan ME, Brihuega B, Pettinari MJ, Cairó F (2011) Multiple-locus variable-number tandem repeat analysis of reference strains used for the diagnosis of leptospirosis in Argentina. Rev Argent Microbiol 43: 251-255.
- Pavan ME, Cairó F, Pettinari MJ, Samartino L, Brihuega B (2011) Genotyping of Leptospira interrogans strains from Argentina by multiple-locus variable-number tándem repeat analysis (MLVA). Comp Immunol Microbiol Infect Dis 34: 135-141.
- Grune Loffler S, Passaro D, Samartino L, Soncini A, Romero G, et al. (2014) Genotypes of Leptospira spp. strains isolated from dogs in Buenos Aires, Argentina. Rev Argent Microbiol 46: 201-204.
- Grune S (2014) Isolation and genotypic characterization of leptospires from wild animals in three Argentinean ecoregions using the Multiple-Locus technique Variable-number tandem repeat Analysis (MLVA): Coincidence with genotypes from production animals.
- Gower JC (1996) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53: 325-338.
- Scialfa E, Brihuega B, Venzano A, Morris WE, Bolpe J, et al. (2013) First Isolation of Leptospira interrogans from Lycalopex griseus (South American Gray Fox) in Argentina Shows New MLVA Genotype. J Wild Dis 49: 168-172.
- Campagna L, Benites P, Lougheed SC, Lijtmaer DA, Di Giacomo AS, et al. (2011) Rapid phenotypic evolution during incipient speciation in a continental avian radiation. Proc Royal Soc Biol Sci.
- He Y, Caporaso JG, Jiang XT, Sheng HF, Huse SM, et al. (2015) Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome 3: 34.
- Zuerner RL, Alt DT (2009) Variable nucleotide tandem-repeat analysis revealing a unique group of Leptospira interrogans serovars Pomona isolates associated with California sea lions. J Clin Microbiol 47: 1202-1205.
- Koizumi N, Izumiya H, Mu J, Arent Z, Okano S, et al. (2015) Multiple-locus variable-number tandem repeat analysis of Leptospira interrogans and Leptospira borgpetersenii isolated from small feral and wild animals from East Asia. Infect Genet Evol 36: 434-440.
- Koizumi N, Muto MM, Izumiya H, Suzuki M, Ohnishi M (2015) Multiple-locus variable –number tandem repeat analysis and clinical characterization of Leptospira interrogans canine isolates. J Med Microbiol 64: 288-294.
- Zilber AL, Picardeau M, Ayral F, Artois M, Demont P, et al. (2014) High-Resolution Typing of Leptospira interrogans strains by Multispacer Sequene Typing. J Clin Microbiol 52: 564-571.
- Chideroli RT, Pereira UP, Gonçalves DD, Nakamura AY, Alfieri AA, et al. (2016) Isolation and molecular characterization of Leptospira borgpetersenii serovar Hardjo strain Hardjobovis in the urine of naturally infected cattle in Brazil. Genet Mol Res 15.
- Slack AT, Dohnt MF, Smythe LD (2005) Development of a multi-locus variable number tandem repeat analysis (MLVA) for Leptospira interrogans and its application to Leptospira interrogans serovar Australis isolates from Far North Queensland, Australia. Ann Clin Microbiol Antimicrob 4: 10.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences