In Silico Molecular Interaction Studies of Drugs with Human Serum Albumin-An Overview
Anu Radha Pathania* and Mamta Devi
Department of Chemistry, University Institute of Sciences, Chandigarh University, Gharuan-140413, Mohali, Punjab, India
- *Corresponding Author:
- Anu Radha Pathania
Department of Chemistry
University Institute of Sciences
Chandigarh University
Gharuan-140413, Mohali
Punjab
India
E-mail: anuradha.appsci@cumail.in
Received date: August 09, 2023, Manuscript No. IPMSC-23-17671; Editor assigned date: August 11, 2023, PreQC No. IPMSC-23-17671 (PQ); Reviewed date: August 25, 2023, QC No. IPMSC-23-17671; Revised date: February 03, 2025, Manuscript No. IPMSC-23-17671 (R); Published date: February 10, 2025, DOI: 10.36648/IPMSC.9.1.001
Citation: Pathania AR, Devi M (2025) In Silico Molecular Interaction Studies of Drugs with Human Serum Albumin-An Overview. J Mol Sci Vol:9 No: 1
Abstract
The main focus of this paper is the use of various computational techniques and the experimental applications of computational techniques for studying the interactions of proteins with various drugs including both quantitative and qualitative aspects. Since the period of ten years, proteins are considered as the cellular target for many cytotoxic anticancer agents and now putting an eye on how various drug molecules going to interact with various proteins followed by their applications. Beyond this their advantages in the medical field for drug delivery and development are also discussed. However, few other techniques such as fluorescence and absorption spectroscopy were also put in use while studying drug-protein interactions and the docking results allow us to cross-check the modes of interaction of ligands with particular proteins. Along with this it assists to find their estimated binding affinities with target protein. Moreover, the field of molecular docking has become an area of interest among various other research branches like medicine, molecular biology, chemistry, and so on. Besides the applications and mechanisms of docking, various methods of effective and specific drug design are also enlisted and this is done to lower down their side effects and make them more efficient. Furthermore, the study of various chemical and conformational modifications that are caused by these drug-protein interactions are also discussed. Hence by using more computational methods for analysing drug-protein interactions could be the basis of future innovations in manufacturing more efficient and pocket friendly drugs.
Keywords
Molecular docking; Molecular interactions; Proteins; Drugs; Computational methods
Introduction
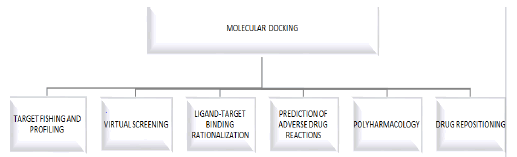
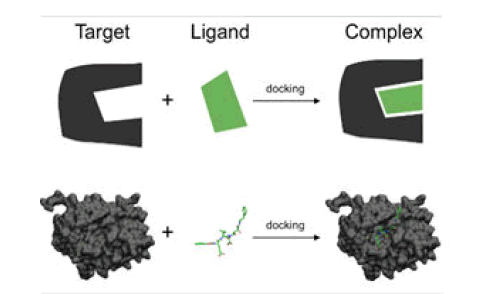
Molecular docking made its first appearance in the mid of 1970 and also it works as a unique in silico tool in drug discovery and design. In the last decades, in silico approaches are used in various fields such as chemoinformatic, artificial intelligence, and molecular modeling [1]. Moreover, molecular docking is the most successful structure based on computational techniques. It predicts the interaction that takes place among different biological targets and drugs [2]. From the early 1980’s, one of the studies of interest regarding the utilization of molecular docking in drug discovery and its biological target could be noted. This study includes the computational method that enables the geometrical structures of ligand-receptor alignments which are feasible that is for the structure of thyroxine and heme-myoglobin. Since then, the applications of docking are kept on enhancing and today it is widely used in many different tasks related to drug discovery programs [3]. Process included in molecular docking are displayed in Figure 1.
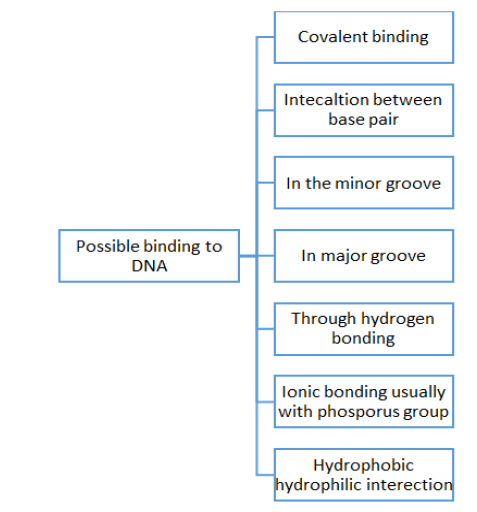
Besides drug designing, but molecular docking is also used in the prediction and simulation of complexes. It also tells regarding the involved mechanism which includes the formation of complexes even at the basic level is molecular one. Molecular docking is a very promising technology in displaying 3-D structures, and understanding the process of formation of drugs and complexes. Molecular docking is based on the principle of "key and lock" pairing [4]. Here in this review, different kinds of drug-protein interactions are discussed that as DNA-drug interaction, HSA-drug interaction, BSA-drug interaction, etc. The technique of molecular docking is successfully developed for investigating better interaction of proteins with small molecules or even for molecules at the atomic level [5]. Besides this, a few other techniques are also known such as fluorescence, equilibrium dialysis, potentiometry, CD spectroscopy, UV-Vis, fluorescence spectroscopy, FTIR, etc., [6] but out of all known techniques to date docking is one of the most successful techniques out of rest techniques used. Also, the approach of molecular docking can be used to find out the activity of any drug [7]. Based on binding energies, firstly the docking score for any drug can be determined. This docking score further helps to find out comparative results for different drugs while interacting with the receptor that is protein [8]. Furthermore, this computer-based in silico approach is not only providing better options for drug delivery but also cuts down the cost of development of drugs significantly [9]. Due to this reason, the silico-based screening of various compounds against various biological targets is gaining attention and also meets challenges such as the discovery of more efficient antiviral drugs. Also, this drug-protein interaction is known to be one of the most crucial pharmacological processes which not only affects the structure but also the physiological process of the carrier protein. Besides this, it also affects the elimination and distribution of various drugs [10]. Also, the efficiency of any drug is directly related to the degree of binding and drug-protein affinity. Moreover, this molecular docking technique helps to reveal the binding location of various proteins with the binding interaction of any particular drug [11]. For a better understanding of the concept of molecular docking in drug-protein interaction let's take an example of protein-DNA interaction with drugs using computational studies. In the case of DNA, the binding depends on the condition of nature of metal and ligand only than various drugs tends to bind to different sites of DNA and all possible bindings are displayed in Figure 2 [12].
Materials and Methods
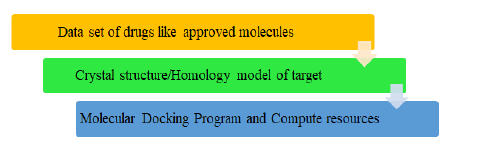
Beyond molecular docking, DNA binding capabilities could be investigated by a few other techniques such as fluorescence, electrochemical, adsorption, hydrodynamic studies, etc. But out of all these, molecular modeling proved to be the most successful technique. For instance, the antimicrobial activity of a ligand was tested using Cu (II) complex against the walls of strains of bacteria using molecular docking. In this way, by using a hemolytic assay on human red blood cells the toxic nature of the Cu (II) complex was observed [13]. Also, the concluding results that were observed using docking techniques were much clearer than others. To interact in a better way a drug must fulfill the following requirements. It should have good solubility in water to enhance its intake at the cellular level, high binding energy with any protein, and high cytosis activity which made it capable of inhibition of protein synthesis of diseased cells, for selective recognition and cleavage there must be specific binding domain and molecular docking takes place by intramolecular hydrogen bonds. Another protein to be considered is HSA which is a non-glycosylated polypeptide protein. Also, HSA is a single chain and greatly solvable plasma protein. A total of 585 amino acids are present in the protein named HSA whose molecular mass is around 66.5 kDa. Also, it possesses a helical structure that lacks any beta sheet particles. Moreover, the drug-HSA binding is the main way of learning the pharmacokinetics of drugs and other effects related to it. For example, anti-inflammatory drugs bind with HSA, and hence its role is quite crucial in drug transportation, distribution, elimination, and delivery of the drug. Another case study reveals that in the DF-Na-HSA binding is the aquaphobic interaction where the role played by HSA is the crucial one. The mode which is involved in the binding of the groove is found to be more successful for the drug-HSA interaction. Such results would help in developing their potential biological, physiological implications and pharmaceutical implications. Also, to manage the COVID pandemic a lot of efforts are done by scientists globally. For new drug discovery, few of the existing drugs were rerouted using molecular docking for saving time and handle the situation wisely. As a result, a lot of drug activities were evaluated using virtual screening to cure the widespread of the novel coronavirus. The process of virtual screening is given in Figure 3.
Further for a better understanding of the concept of molecular docking in drug-protein interaction followed by applications of molecular docking with a few detailed examples are discussed.
Basics of molecular docking
Molecular docking is also known as computer-aided drug design which is conventionally used for drug invention and designing. Usually, the drug interacts with the target in a noncovalent manner. Consequently limiting the inventions and research. Nowadays there is a lot of molecular docking software which are of higher calculation and predicts results more accurately [2]. Moreover, docking between two molecules could take place in two ways namely through non-covalent or covalent interactions. Generally, non-covalent interaction includes Van Der Waal interactions, electrostatic interaction, and hydrogen bonding. Also, it uses knowledge based scoring functions to characterize such interactions. Besides this, drugs bind through covalent interactions as well to active sites of the protein (Table 1). Out of these two non-covalent interactions are mainly used. Additionally, the basic difference between these two interactions is listed in Figure 4.
|
S. no. |
Covalent interactions | Non-covalent interactions |
|---|---|---|
|
1 |
Off-target reactivity and toxic | Targeted reactivity and less toxic |
|
2 |
Leads to the development of highly selective inhibitors. | Inhibitors developed are less selective |
|
3 |
Novel inhibitors with increased potency are developed. | Comparatively lower potency. |
|
4 |
Able to target extremely small molecules such as propyl dipeptidase which is used to treat human brain disorder. | Unable to target extremely small molecules. |
|
5 |
Extensive computational cost. | It is pocket friendly. |
|
6 |
Software used-covalent dock Example: A drug named curcumin which can be obtained from plants named curcuma londa that is usually found in eastern parts of India. |
Software used-SAMSON Example: Drugs named amprenavir, indinavir, anagliptin etc interacts through non-covalent interactions. |
Table 1: Difference in covalent and non-covalent interactions.
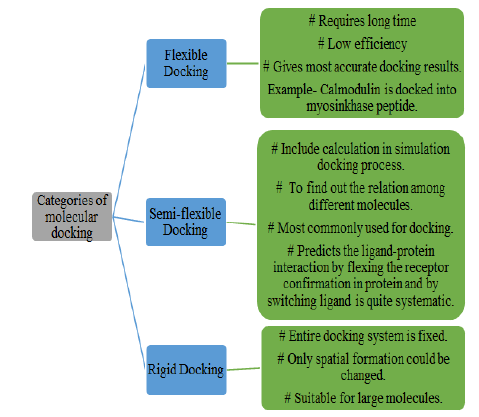
Also, docking has three different parts namely-flexible docking, semi-flexible docking, and rigid docking. A detailed description of all these three categories is enlisted in Figure 4.
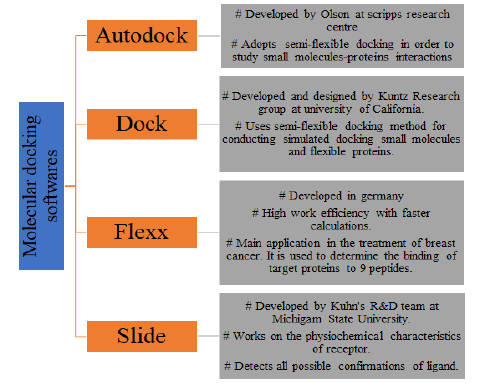
Nowadays, a lot of software’s are there which can be used for the purpose of molecular docking and give better results in stipulated time. Four mainly used software’s for docking along with their brief description are given in Figure 5.
Results and Discussion
Application of molecular docking in studying drugprotein interaction
To study BSA-drug interaction: Bovine Serum Albumin (BSA) is a plasma protein that contains a total of 583 amino acid residues that bind through a single chain. In this particular investigation, to understand the glutathione-BSA interactions, a software named Argus Lab software was used as a computational dating program.
Moreover, for the sake of finding the favored site for binding and mode of glutathione when docked with a protein named BSA and the molecular docking results were complemented by the results of both fluorescence and UV-Vis spectroscopic techniques. The BSA protein has two drug binding grooves namely binding/ Sudlow’s site 1 and 2. These binding grooves are there in aquaphobic areas of subdomain 2A and 3A and glutathione ties up with site 1 of subdomain 2A pocket in domain 2 of BSA protein. Moreover, glutathione tends to go for hydrogen bonding with residues. Also, the surroundings of glutathione possess a total of three amino acid residuals of three different binding areas 8.99 kcal/mol was the resulting binding energy for the complex and with the help of these docking results obtained after docking the complex is formed after glutathione-BSA interactions. Additionally, the main point to be noted is that different aquaphobic and aquaphobic acids are connected with glutathione when the required complex is formed. Hence, as a result, molecular docking reveals an indestructible interaction among both that is the ligand and the protein.
The reason behind this is the formation of hydrogen bonds in the required complex. Moreover, these results show similar results as shown by fluorescence studies as well. The reason behind such strong interaction among the complex formed by glutathione and BSA could be explained based on the presence of three amino acids and the big chemical structure of the compound. Another ligand named NFLX was docked with BSA by using B3LYP/6-311G. One of the best conformers of the protein was docked to discover the binding site of BSA. Besides the binding sites, it also helps to determine interactions between NFLX-BSA and as a result, two hydrogen bonds tend to form between the two and are between the hydroxyl groups of NFLX. Hence, different sites of NFLX bind with different sites of the binding protein. Also, the least free energy of the BSA-NFLX was found to be 3.69*105 Lmol-1 and -29.63 KJ/mol respectively. Moreover, these docking results were found to be almost similar to that the results obtained experimentally.
Molecular docking in drug-DNA interaction: To understand drug-DNA interactions, molecular docking proved to be a very useful and attractive tool that helps in drug design and discovery for future developments. Molecular docking also predicts the structure and orientation of ligands inside the DNA groove and their binding capability. Moreover, according to docking results, ligand (DEEI) and Cu (II) complex bind within a minor groove of protein that is DNA efficiently. The binding energy for ligand (DEEI) is 302.6 kcal/mol and for another complex is 374 kcal/ mol. The reason behind this is that Cu (II) complex is having more negative binding energy due to the formation of stronger Cu (II)-DNA interactions. Another case study of molecular modeling in the case of drug-DNA interaction is the member of the third generation is Lomefloxixacin (LMF) in which quinoline antibiotics are fluorinated and have a piperazinyl moiety both at position 6 and 7 respectively. It is capable of penetrating through the cell walls. This LMF is used on the large scale for the treatment of many infections like soft tissue infection, typhoid, bone and joint infections, and so on. Moreover, the role of molecular modeling is vital in drug designing and discovering new drugs. Also, it was observed from the observations that the binding sites of LMF were present in the groove of ctDNA.
Furthermore, the docking simulation helps in calculating the change in free energy (ΔG°) of binding force ctDNA-LMF which was -23.73 KJ/mol. This shows that the LMF interacts with ctDNA through the minor binding groove. Moreover, the data collected regarding free binding energy from fluorescent emission are according to the results estimated by using docking analysis but the resulting values of binding energy which are calculated by using molecular modeling and fluorescence emission experiment are not similar. This is due to the difference in the experimental condition of the two techniques.
Pd (II)-DNA and Pd (II)-BSA interactions: Along with the foretelling of interaction among the molecules and macromolecules, molecular docking could be applied to detect the binding character and orientation of the molecule. As a result of which the resulting complex is formed with the least total energy. Moreover, the outcomes of various experiments conducted are flattered by molecular modeling where the Pd (II) complex is docked to both of the proteins that are DNA and BSA. This was done to highlight the binding modes and sites. Also, the free binding free energy of the complex (Pd(acac)(Val)) is quite less in comparison to (Pd(acac)(Ala)) which results that (Pd(acac) (Val)) showing a hike in binding ability in comparison to (Pd(acac)(Ala)) ligand. Moreover, the molecular docking of the complex with DNA depicts that both of the ligands are present in the major groove region of DNA and these findings are quite similar to that of the observations obtained experimentally. Furthermore, to learn the consequences of any drug molecule which is a biologically active molecule, the location of binding in the model transport protein environment should be known. Also, it can be observed that there are three homologous domains inside BSA which is a globular protein that is I, II, and III which are further categorized into nine loops by seventeen disulfide bonds. As a result of this, each domain is formed of two sub-domains that are A and B. Also, the main region inside the hydrophobic pockets of the ligand tends to bind with BSA. These aquaphobic pockets in two sub-domains IIA and IIIA are named by the site I and site II. Hence, both of these compounds delineated the capability of effective binding with both of the proteins that are DNA and BSA. Also, this is worth studying DNA and BSA interaction with the above-mentioned complexes as it includes detailed information on the anti-cancer and antiinfective activity of the two above-mentioned complexes.
Drug-human serum albumin interactions: HSA has a total of 585 amino acids and also is composed of three homologous domains in a structure that is- I, II, and III. In this I consists of 1-195 amino acid residues, II have 196-383 and 384-585 are there in III domain. HSA is a protein with higher stability and 3D structure. Also, its structure is susceptible to various environmental factors including ionic strength and pH levels. For inspecting the binding of tiny ions with HSA following methods can be used.
• Fluorescence
• Synchronous fluorescence
• UV visible
• Equilibrium dialysis
• Potentiometry
• CD spectroscopy
• Docking
Here we will discuss the drug-HSA interaction using molecular docking. The main area of binding of ligand to HSA lies in the aquaphobic pockets in subdomains II and III which relates to site I and site II here to find out the binding mode of CBDA to HSA, the Autodock 4.2 program was used. Here, CBDA gets placed into an aquaphobic pocket of the protein. Moreover, the CBDAHSA complex was balanced by the formation of hydrogen bonds between drug, protein, and some amino acid residues. Additionally, the stability of binding energy scores of drugprotein interactions is calculated by using molecular docking [7]. When FLV interacts with HSA, studies related to molecular docking were conducted in contemplation to study the interaction of FLV with HSA. As per 2D and 3D poses, FLV forms an interaction that is strong hydrophobic in nature and p-p stacking with Trp-214. Although, a good docking score of about 9.623 was observed. Here, FLV tends to bind at sub-domain IIA at the site I of HSA. Additionally, the main forces behind the FLVHSA binding interaction were hydrophobic interactions. Now, on putting an eye on the binding of DHP with HSA. DHP interacts with both the binding sites of the HSA molecule which are site I and site II. It interacts with hydrogen bond formation and pihydrogen bonding. However, site I of subdomain II A was found to be the major binding on HSA, which is in accordance with experimentally determined values.
Application of molecular docking in drug-discovery for COVID-19
Few of the techniques put forward by many scientists offer more productive results but the outcomes of computational analysis by the use of molecular docking are one of the most promising techniques that are put in use to cure the consequences of COVID-19. In recent studies molecular do king of FDA approved drug is being done with the main protease of SARS-CoV-2 few of the drugs with more efficient binding energy fitting store and non-covalent interactions with receptor are Simeprevir, Ergotamine, Bromocriptine, and Tadalafil. His 41 and cys 145 were the key active site residues non-covalent interaction include hydrogen bonding hydrophobia interactions pi-sulpha and pi-pi interactions. Moreover, Coronavirus genomes are divided into two different groups when it comes inside the host cell. Those two groups are structural and non-structural proteins. During the replication and transcription of Coronavirus, RNA synthesis occurs at cytoplasmic membranes. In converting poly protein into functional pieces, the main protease plays a key role. Additionally, the crystallized structure of the main protease has two chains namely A and B where chain A is used for macromolecule preparation and 306 residues are composed by the protein chain. Few other techniques including molecular docking were put into execution to evaluate the action of drugs and around 1615 FDA-approved drugs were put into consideration using docking software named Autodock Vina for virtual screening. Besides the drug screening by Autodock Vina, the GOLD suit was also put in use to evaluate the fitness score using CHEMPLP as higher the fitness score than there will be better docking interaction between the drug and protein. Also, this Autodock Vina is used to detect the non-covalent interaction between drug and protein. Besides this, a lot of molecular docking calculations were performed. These calculations were done between the target protein and some of the selected ligands. Moreover, it was found that CID22674959 and CID89869520 have no activity against the formation of a few of the compounds like favipiravir, such compounds are found to be active in all kinds of formations. Also, favipiravir interacts with 5W44, 6FS8, and 6QX8 whereas the basic structure of favipiravir was found to hinder the protein interaction. Further, the criteria that are put under consideration for the identification of better drugs are docking score and total energy where the docking score of any drug/ligand must be good in comparison to the substance taken for reference. However, in a few studies, it was found that 2-oxo-1H-pyrazine-3-carboxamide gives better docking results as compared to favipiravir. Besides molecular docking, the better performance of the former compound is proved by molecular mechanics Poisson-Boltzmann surface area and analysis of ADME. When molecular docking was done between the selected drugs with specific protein targets then CID22674959 and CID89869520 were found to be inactive against RdRp protein targets. A series of calculations were performed to get better results. Another ligand named Nglycon was considered to interact with SARS-CoV-2 where Nglycon was found to be fitted completely inside the pockets of SARS-CoV-2 spike protein where binding occurs. It forms one hydrogen bond with Asn90 and a few other formations were observed with better binding interactions and modes as well. Moreover, variable electrostatic bonds were used to stabilize the site of binding for spike protein [5], and the process involved is shown in Figure 6.
One more drug named Rutin was identified as an active path and phytochemical against many proteins by using molecular docking [8]. Moreover, Rutin tends to inhibit both structural and non-structural proteins. Based on various literature and available crystal structures, the recognition of critical remnant of binding pockets and mechanism that is involved in inhibition of residues were analyzed. The binding energy and structure of Rutin were found to be favorable to binding with the target molecule.
Few of the drugs with their interaction are listed below:
Amoxicillin: It is a penicillin antibiotic that helps in fighting different bacteria and is also useful in the treatment of many diseases. As it does not produce any strand breaks in DNA. Along with this, it insolates the plasmid of DNA which would pierce through the cells. Also, the stimulation of the drug at the cellular level is required to cause any damage to DNA. Moreover, the association of the evolution of crosslinks along with DNA in gastric mucosa cells is with the exertion of reactive oxygen molecules at cellular levels. Also, in accordance with a few studies, the drug tends to induce modification inside DNA that is oxidative base moderation, and this activation is linked with the generation of free radicals. In comparison to non-infected cells, 8-hydroxydeoxyguanosine and single-stranded DNA peaks are higher in H-poly infected gastric mucous cells. Furthermore, the results of the penicillin-binding protein show that free radicals are involved in DNA lesions' formation which was induced by the drug Amoxicillin itself. Amoxicillin protects the DNA damaging activity by preventing it against the potential side effects of eradication and drug-protein interaction is shown with the help of the diagram (Figure 7).
Cephalosporin: In some studies, the interaction of four cephalosporins which include cefalexin, cefaclor, cefirime, and cafepime with protein that is HSA is included. Moreover, the structure of cefalexin has chlorine in the third position and both are second-generation antibiotics. One of the applications of cefaclor is that it is used to cure the infections in respiratory tract along with pathogens that are gram-positive or negative. Additionally, a 4th generation drug cefepime also acts as an antibiotic. Also, it has an effective antimicrobial effect and helps in the treatment of pneumonia, sepsis, etc. On the same note, cafepime occurs through renal function and it tends to cause renal toxicity when consumed in a higher dose. The most abundant transport protein is HSA which is present in blood plasma and also possesses the ability to bind with diverse and large ligands. For further discussion, the molecular docking results are taken from the recently conducted studies. The docking results show the flexible nature of the single bond present in the R-2 group of cafalexin and cefaclor. This results in the binding of HSA to both the sites for the sake of the formation of a ground state complex. However, the carboxylic group present in cefepime tends to form hydrogen bonds at the first site of HAS.
Ceftriaxone and rutin: As per many recent studies, human serum albumin tends to interact with drugs in different ways. Also, the basis of albumin's transport function is its capability of binding with many biologically active structures and medicinal substances. Moreover, the ligand becomes more soluble due to the binding ability of albumin and it can form a protective layer and would protect the compounds which bind through enzymatic degradation and oxidation. Furthermore, the binding ability of albumin makes a remarkable contribution to drug pharmacokinetics. When the technique of docking was used employed to learn drug-HSA interactions that are ceftriaxone and rutin. Molecular docking helps to analyze the binding energy and the structure of the complex. Further, docking was applied on both binding sites of HSA. As per docking calculations, the main contribution to the calculation of free binding energy was done by desolvation energy and Van Der Waal interactions. However, a small contribution of electrostatic interactions and change of configuration energy was also observed. In general, there are two binding sites in HSA where ligands bind with variable affinities. Besides this, a series of analyses were performed for both of the ligands on both of the sites of protein, and for analysis, the densely populated clusters which possess the least free energy values were considered.
Rifampin and isoniazid: These two above listed drugs are used to treat tuberculosis and out of these, isoniazid was discovered in 1952. This was the first drug to cure tuberculosis disease. Moreover, isoniazid opposes the preparation of nojolic which was the reason behind the formation of bacterial cell walls however this drug has few side effects such as gastrointestinal and epigastric distress vomiting, and pancreatitis. Another one rifampin was introduced in 1967 and is a bactericide antibiotic used to treat tuberculosis itself. As per the findings of the conducted studies regarding the interaction of these drugs with pepsin, HSA, and a few other macromolecules which are imported. Using molecular docking the binding of both drugs with pepsin can be studied. Both of these drugs tend to tie up with the active site of any enzyme where 9.2 kcal/mol and 8.3 kcal/mol are the interaction energies of both isoniazid and rifampin respectively. Out of this two isoniazid tends to interact with pepsin more strongly in comparison to the drug. Due to the presence of more H-bonding the isoniazid to form a stronger complex than rifampin moreover similar results were obtained from both experimental bond molecular hence the binding energy of both drugs in action can be related to the tie up of casein to the enzyme and hence the enzymatic activity of the drugs increases.
Meropenem: The biological target for meropenem is Penicillin Binding Protein (PBP) and it is an anti-infective drug. The drug binds with residues of human serum albumin through electrostatic interactions and these results agree with thermodynamics results. As per studies, electrostatic interactions tend to stabilize HSAmeropenem complex. Although molecular docking also shows similar results that is hydrogen bonding and some interactions can be observed in meropenem-HSA interaction. As per studies done recently, it was found that the crystal formation of meropenem bond to L, -D-Transpeptidase of Mycobacterium tuberculosis using hydrogen bonding and some of the aquaphobic interactions and consequently leave the dynamic site of the protein blocked. Later Lys-436 of HSA was replaced by Ala-436 this was done to alter the active site of any protein and Meropenem. Hence this mutation fully transposed the mode of meropenem-HAS binding near Sudlow's site II and was bind bear IB site. Hence a major role in binding near Sudlow's site II and Lys-436 in the stabilization of the HSA-meropenem complex.
Aspirin/Diflunisal: Both of these drugs are from the salicylate category of non-steroidal and anti-inflammatory drugs. They are having large pharmacological and biological functions and their bonding with the protein that is DNA offers insight for rational designing and development of effective therapeutic agents. Also, to find out the way of binding drug-DNA the technique of molecular docking was put in use. This tool help in finding the affinity of binding of a particular ligand with its respective receptor which relates with experimentally calculated values as well. In this case, rigid molecular docking was carried out. This is done to construe the binding mole of diflunisal and aspirin with DNA complex. Their observed binding sequence is a (CGCGATTCGCH) 2 dodecamer (PDBID:18non). Moreover, the ligand taken is flexible this is done to obtain variable confirmation and the analysis of their most favourable docked position was done. Additionally, the energy of binding of aspirin was found to be around -4.71 kcal/mol and its GC-rich region was revealed by docking results. The possibility of hydrogen bonding in aspirin was revealed by groups of aspirin having oxygen (01) which are present in the proximity of deoxyguanosine in DG10 of chain A of dodecamer. In addition, diflunisal is having a binding energy of -3.65 kcal/mol and in this, the formation of hydrogen bond was revealed by docking studies and binding takes place between oxygen bearing group and 8th number thymine of single strand of DNA. In this oxygen atom and a main severe for hydrogen bond receptor.
Diclofenac: Under simulative physiological conditions, an experiment was conducted to learn Diclofenac Sodium (DF-NA)- Human Serum Albumin (HSA) interactions using a molecular modeling technique. This particular technique can provide insight into the interaction between ligands and macromolecules. Moreover, molecular modeling has wide application in the designing and development of new drugs. As per results from recent the location of DF-Na-HSA was revealed that is inside the minor groove of the protein and there is hydrogen interaction of C-2 amidogen H21 N3 and O-40 of G/6 with carbonyl 0 atoms. Besides carbonyl-0 atom it interacts with hydroxyl 0 and amidogen H of DF-NA as well. The hydrogen bond length of these three was observed as 2.1, 2.0, and 2.4 respectively hence the use of hydrogen bond was revealed by these results. Moreover, then docking results are found to be complements by experimental results obtained from other techniques lastly the combination of both of the results could be put in use to study the various interaction of smell molecules and biomacromolecules which provide information about different aspects in detail.
Few other drugs with their scores of dockings and sites of binding are listed in the Tables 2 and 3 below.
| S. no. | Name of drug/ligand | Docking score | Binding site |
|---|---|---|---|
| 1 | Remdesivir | 5.9 | RdRp and Mpro |
| 2 | Fluconazole | -7.3 | Rhizopus lanosterol 14-alpha demethylase |
| 3 | Isavuconazole | -9.4 | Rhizopus lanosterol 14-alpha demethylase |
| 4 | Pramiconazole | -11.8 | Rhizopus lanosterol 14-alpha demethylase |
| 5 | Itraconazole | -11.6 | Rhizopus lanosterol 14-alpha demethylase |
| 6 | Posaconazole | -11.1 | Rhizopus lanosterol 14-alpha demethylase |
| 7 | Ketoconazole | -10.6 | Rhizopus lanosterol 14-alpha demethylase |
Table 2: Drugs and their docking scores with binding sites.
| S. no | Name of drug/ligand |
Docking score | Role/Use |
|---|---|---|---|
| 1 | Piroxican | -9.70 | To treat pain, tenderness and stiffness. |
| 2 | Tolmatin | -7.60 | To treat mild pain and arthritis. |
| 3 | Celecoxib | -9.10 | To treat symptoms of arthritis. |
| 4 | Penazopyridine | -7.10 | To treat pain in urinary tract. |
| 5 | Mefenamic acid | -7.60 | To treat minor pain. |
| 6 | Methyl Salicylate | -6.20 | To treat aches and joint pain. |
| 7 | Omnicef | -9.87 | To treat bacterial infection. |
| 8 | Disulfiram | -3.15 | Proteasome |
| 9 | Pexidartinib | -6.24 | PK inhibitor |
| 10 | Amperenavir | -5.50 | Protease inhibitor |
| 11 | Vilantecol | -5.78 | Adrenergic receptor modulators |
| 12 | Edoxabon | -7.97 | AEC inhibitors |
| 13 | Acotiamide | -8.07 | To treat function dyspepsia |
| 14 | Rivaroxaban | -5.26 | AEC inhibitors |
| 15 | Cilazaprilat | -5.79 | Direct oral anti -coagulants |
| 16 | Arformoterol | -5.61 | Adrenergic receptor modulators |
| 17 | Dasantinib | -10.46 | To treat antiviral activities and MERS-COV |
| 18 | Indinavir | -6.31 | Protease inhibitor |
| 19 | Copanlisib | -6.90 | PK inhibitor |
| 20 | Bentiromide | -8.80 | Used in screening test for exocrine pancreatic insufficiency. |
| 21 | Lymecycline | -6.78 | To treat acne and bacterial infection |
| 22 | Canagliflozin | -5.87 | To treat high blood sugar |
| 23 | Anagliptin | -6.08 | Protease inhibitor |
| 24 | Darolutamide | -10.03 | To treat prostate cancer |
| 25 | Lafutidine | -8.20 | To treat peptic ulcer and gastroesophageal reflux disease. |
| 26 | Boceprevir | -6.01 | Protease inhibitor |
| 27 | Vilazodone | -5.98 | To treat depression |
| 28 | Semagacestat | -9.36 | Protease inhibitor |
| 29 | Methotrexate | -5.41 | To treat cancer |
| 30. | Dutasteride | -5.91 | Primary proteinase inhibitor used to treat prostate cancer. |
Table 3: Drugs and their docking scores with uses
From Table 3, it could be noted that lamofloxacin is having a higher negative docking score (-23.73). Also, a more negative docking score denotes the strong binding of the drug with the particular target protein. Hence, lamofloxacin binds strongly with the target protein which is DNA. Furthermore, Imatinib also binds strongly with the target protein (having a docking score of -20.04). Few other drugs such as fluconazole, isavuconazole, pramiconazole, itraconazole, posaconazole, ketoconazole, albaconazole, ravuconazole, pyrozoline, isoquercetin, vepaol, nimbidiol, vilasinim, nimbocinolide, xaempferol, isonimouinolide and banbaloin are having almost similar docking scores which are -7.3, -9.4, -11.8, -11.6, -11.1, -10.6, -9.3, -9.2, -6.2, -9.73, -9.43, -6.8, -6.62, -6.55, -6.44, -6.29 and 7.54 respectively. However, remdesivir is having the least docking score (5.9) out of all drugs. Besides all these some other drugs are also mentioned along with their docking scores and their uses to treat different diseases. A drug named dasantinib is found to have the most negative docking score which is -10.46 and is also used for COVID-19 treatment. Rest all drugs lies between the docking score 5-10 that is piroxican, tolmatin, celecoxib, penazopyridine, mefenamic acid, methyl salicylate, omnicef, pexidartinib, amperenavir, vilantecol, edoxabon, acotiamide, rivaroxaban, cilazaprilat, arformoterol, indinavir, copanlisib, bentiromide, lymecycline, canagliflozin, anagliptin, darolutamide, lafutidine, boceprevir, vilazodine, semagacestat, methotrexate and dutasteride with docking score -9.70, -7.60, -9.10, -7.10, -7.60, -6.20, -9.87, -6.24, -5.50, -5.78, -7.97, -8.07, -5.26, -5.79, -5.61, -6.31, -6.90, -8.80, -6.78, -5.87, -6.08, -10.03, -8.20, -6.01, -5.98, -9.36, -5.41 and -5.91 respectively. But disulfiram is having the least docking score out of all the above-mentioned drugs and will not bind strongly to the target protein.
Conclusion
With the help of docking, one could easily find out and verify various factors such as various experimentally performed tasks and various effects which include solvent, entropy and enthalpy effects, binding energy, and so on. This technique is pocket friendly in comparison to others and hence can be applied for virtual screening of the targeted protein. After the mid of nineteenth century, molecular docking is found to be a useful tool in drug discovery and development. The results of molecular docking are based on quantum mechanics Moreover, considering the availability of a lot of in silico software and methods. Also, there are uncountable possibilities for new developments and innovations. Molecular docking has very vast applications in the fields of drug repositioning, drug discovery, prediction regarding after effects of drugs, or even in the fields of software development. As discussed, docking results would predict the binding of any protein with the respective drug and besides this, it also provides information regarding binding sites and structures of the compounds or drugs. Also, studies show that in most of the cases docking results were found to be almost equivalent to the results obtained from MD simulations which proves the success of molecular docking in the field of drug discovery and development. The application of ligand profiling and repositioning is expected to provide Novel opportunities in the field of molecular docking for drug delivery and development. Hence, this technique provides better results in stipulated time and also at lower costs and it is also believed that the coordination of molecular docking with computational analysis would bring promising developments in the field of pharmacy for designing various drugs and would ultimately help in the improvement of already known drugs in order to enhance their activity and selectivity against the infected sites.
References
- Pinzi L, Rastelli G (2019) Molecular docking: Shifting paradigms in drug discovery. Int J Mol Sci 20:4331
[Crossref] [Google Scholar] [PubMed]
- Alonso H, Bliznyuk AA, Gready JE (2006) Combining docking and molecular dynamic simulations in drug design. Med Res Rev 26:531-568
[Crossref] [Google Scholar] [PubMed]
- Salmaso V, Moro S (2018) Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front Pharmacol 9:393738
[Crossref] [Google Scholar] [PubMed]
- Li T, Guo R, Zong Q, Ling G (2022) Application of molecular docking in elaborating molecular mechanisms and interactions of supramolecular cyclodextrin. Carbohydr Polym 276:118644
[Crossref] [Google Scholar] [PubMed]
- Al-Karmalawy AA, Dahab MA, Metwaly AM, Elhady SS, Elkaeed EB, et al. (2021) Molecular docking and dynamics simulation revealed the potential inhibitory activity of ACEIs against SARS-CoV-2 targeting the h ACE2 receptor. Front Chem 9:661230
[Crossref] [Google Scholar] [PubMed]
- Vique-Sanchez JL (2021) Potential inhibitors interacting in Neuropilin-1 to develop an adjuvant drug against COVID-19, by molecular docking. Bioorg Med Chem 33:116040
[Crossref] [Google Scholar] [PubMed]
- Mhatre S, Patravale V (2021) Drug repurposing of triazoles against mucormycosis using molecular docking: A short communication. Comput Biol Med 136:104722
[Crossref] [Google Scholar] [PubMed]
- Rahman F, Tabrez S, Ali R, Alqahtani AS, Ahmed MZ, et al. (2021) Molecular docking analysis of rutin reveals possible inhibition of SARS-CoV-2 vital proteins. J Tradit Complement Med 11:173-179
[Crossref] [Google Scholar] [PubMed]
- Konappa N, Udayashankar AC, Krishnamurthy S, Pradeep CK, Chowdappa S, et al. (2020) GC–MS analysis of phyto constituents from Amomum nilgiricum and molecular docking interactions of bioactive serverogenin acetate with target proteins. Sci Rep 10:16438
[Crossref] [Google Scholar] [PubMed]
- Lu XG, Wang Z, Cui Y, Jin Z (2014) Computational thermodynamics, computational kinetics, and materials design. Chin Sci Bull 59:1662-1671
- Kumalo HM, Bhakat S, Soliman ME (2015) Theory and applications of covalent docking in drug discovery: Merits and pitfalls. Mol 20:1984-2000
[Crossref] [Google Scholar] [PubMed]
- Rasool F, Khalid M, Yar M, Ayub K, Tariq M, et al. (2021) Facile synthesis, DNA binding, urease inhibition, anti-oxidant, molecular docking and DFT studies of 3-(3-Bromo-phenyl)-1-(2-trifluoromethyl-phenyl)-propenone and 3-(3-Bromo-5 chloro-phenyl)-1-(2-trifluoromethyl-phenyl)-propenone. J Mol Liq 336:116302
- Almutairi FM, Ajmal MR, Siddiqi MK, Amir M, Khan RH (2020) Multi-spectroscopic and molecular docking technique study of the azelastine interaction with human serum albumin. J Mol Struct 1201:127147
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences