ISSN : 2634-7164
Journal of Medical Microbiology and Immunology Research
Avirulent Gram-negative Bacteria E. coli K-12 or E. coli C Compared with Gram-positive Virulent Diplococcic Streptoccocus pneumonia
Sunil Palchaudhuri1*, Hansini Mundigala1, Eldar Kurtovic1, Anubha Palchaudhuri2 and Archita Biswas2
1Department of Immunology and Microbiology, Wayne State University School of Medicine, 540 E Canfield St, Detroit, MI 48201, USA
2Atlanta Health Centre, Kolkata, India
- *Corresponding Author:
- Sunil Palchaudhuri
Department of Immunology and Microbiology
Wayne State University School of Medicine
540 E Canfield St. Detroit, MI 48201, USA
Tel: +13135771335
E-mail: spalchau@med.wayne.edu
Received date: November 24, 2017; Accepted date: December 07, 2017; Published date: December 18, 2017
Citation: Palchaudhuri S, Mundigala H, Kurtovic E, Palchaudhuri A, Biswas A (2017) Avirulent Gram-negative Bacteria E. coli K-12 or E. coli C Compared with Gram-positive Virulent Diplococcic Streptoccocus pneumoniae. J Med Microbiol Immunol Res 1:3.
Copyright: © 2017 Palchaudhuri S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Growth curve of E. coli C is similar to that of the pathogenic Gram positive strain Streptococcus pneumoniae (Sp). In 1928 Dr. Griffith studied Smooth and Rough colonies on blood agar (BA) medium and significance of his observation has been ignored for a long time. In 1944, Dr. Avery et al. have made wrong interpretation of this observation. TCA insoluble precipitation contained only a few nucleotides or at bests a fragment of 0.1 Kb. It is unlikely to contain any genetic character. Recently, Palchaudhuri et al. revived and redefined Dr. Griffith’s Smooth and Rough colonies. These colonies are actually the growth curve of S. pneumoniae. Our objective is to demonstrate a way of combating the pathogen S. pneumoniae by utilizing the knowledge of growth pattern.
Keywords
Growth curve; Comparison; Xylitol operon; E. coli C; Streptococcus pneumoniae
Introduction
In 1928 Dr. Fred Griffith has introduced the concept of bacterial genetics in his famous pathology notebook. Grampositive growth curve includes two colonies grown on blood agar media [1]. He has seen two types of S. pneumoniae colonies: SMOOTH and ROUGH. Smooth colonies as observed on blood agar media represent the log phase (early, mid and late log) and rough colonies represent the stationary phase (equivalent to the stationary and decline phase of E. coli K-12). Smooth colony is more severe than the Rough colony. We have proven earlier that the Smooth colony represents bacteria in log phase and Rough represents stationary phase [2]. These two colonies represent the bacterial physiological changes. While they are in log phase, we think these bacteria are responsible for the degree of severity of diseases [1,2]. However the same S. pneumomiae appears to be responsible for several diseases like bacterial pneumonia in children and the elderly, paediatric dental caries, otitis media, meningitis and septicaemia (differentiation of the diseases) [3,4]. In recent years investigators have reported that some nontranslational small RNAs are responsible for the differentiation [5].
During 1960-1975, three different strains of E. coli, E. coli K-12, E. coli B and E. coli C have been used worldwide to develop microbial genetics [6,7]. This includes primarily E. coli K-12, bacteriophages (lytic, lysogenic and semi-temperate) and bacterial plasmid DNA (mostly double-stranded covalently closed circle). Recombinant of E. coli K-12 and E. coli C formed in vivo by genetic cross linkage. E. coli HF4714 thus formed shows some interesting characters of parents. Like E. coli C, it forms plaques when infected with bacteriophage ɸX174 and carries genes needed for the utilization of 5-carbon sugar alcohol xylitol. E. coli C is very small in size (~1 μm), K-12 is long (2.5 μm) but the recombinant E. coli HF4714 is more like E. coli K-12 in size. However such knowledge of genetics has been ignored. Around this time Stanford University School of Medicine faculty and their laboratory technicians begin an in vitro gene cloning experiment (1972-1980) [8-10]. In the formation of such recombinant DNAs, several antibiotic resistant determinants are extensively used although the antibiotic resistance has created a crisis in Medicine. What is the difference between genetic determinants and their transposons? Usually bacterial determinant means genetic character but transposons are mobile DNA elements. Mobile DNA elements are not replicons as defined by Jacob and Monod, but they still multiply [11]. Insertion sequence (IS) and transposons (Tn) are both mobile DNA elements [12,13]. The transposons (Tn) are flanked by IS DNA sequences like IS1 with antibiotic resistance characters and/or genes for exotoxin (stx) production, both stable and unstable [14,15].
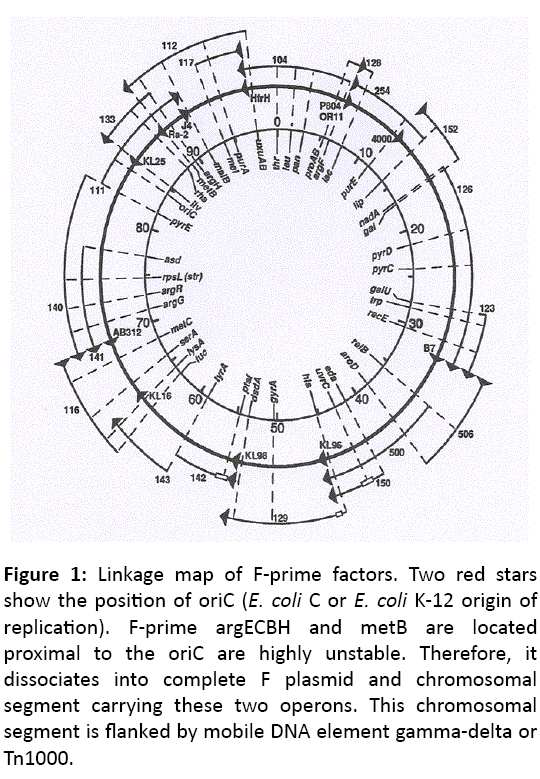
DNA sequence based dissimilarity leads us to conclude that an operon for the metabolism of xylitol (5-carbon sugar alcohol) is absent in E. coli K-12 and E. coli B but present in E. coli C chromosome at 47.5 min location of linkage map (Figure 1), proximal to Met G loci [16,17]. It is well established now that the 10% of E. coli C isolates contain xylitol operon but the origin of xylitol operon remains unknown.
Figure 1: Linkage map of F-prime factors. Two red stars show the position of oriC ( E. coli C or E. coli K-12 origin of replication). F-prime argECBH and metB are located proximal to the oriC are highly unstable. Therefore, it dissociates into complete F plasmid and chromosomal segment carrying these two operons. This chromosomal segment is flanked by mobile DNA element gamma-delta or Tn1000.
Experimental Procedures
All these experiments and bacterial strains have been described in our previous articles [2,14].
Results
Comparison of growth curves E. coli K-12, E. coli C and E. coli HF4714
Growth curves of Gram positive streptococci and Gram negative E. coli K-12.
Previously we have published the growth curve of Grampositive pathogen S. pneumoniae. This belongs to Mitis group streptococci. All members of this group grow in three different phases: pre-competent, competent and post-competent [2]. In many articles investigators have mentioned early, mid and late competent. Linkage map of E. coli C does not differ from E. coli K-12 except for xylitol operon (partial or complete) and therefore the E. coli C is sensitive to single stranded circular bacteriophage ɸX174. However, in E. coli C we have defined log phase in three phases - early, mid and late. Gram-positive S. pneumoniae is also carrying Xylitol operon, but it cannot metabolize except for xylitol phosphate which is highly toxic to their continuity. We have already published how this Grampositive organism is affected by the growth in xylitol (2% or more) [18,19].
Smooth colony of Dr. Fred Griffith represents its growth in log phase (early, mid or late) [1]. Dr. Griffith’s ROUGH colony is the same as stationary phase of E. coli C or E. coli K-12. At the same time, we should not forget that exogenous DNA, single or double, does not have any role in the physiological states (smooth or rough colonies). Meanwhile several distinguished investigators have mixed up the microbiological meaning of a single bacterium with its colony and the bacterial strain (reference not cited). In order to appreciate the difference of colony morphologies, we should go back to the growth curve of E. coli K-12.
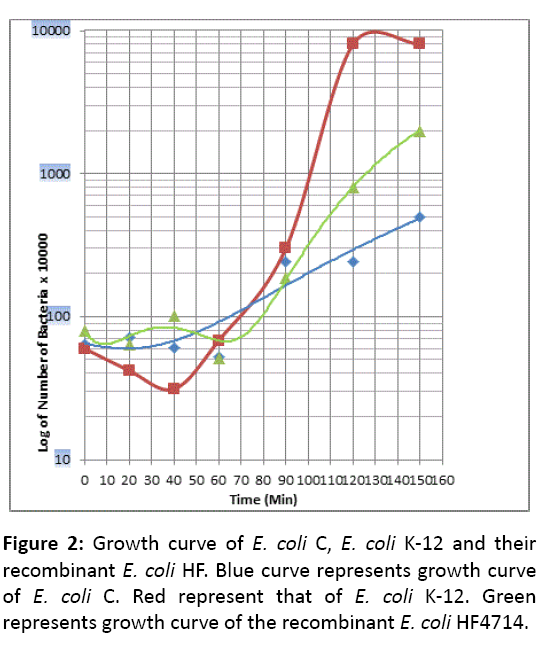
Unlike E. coli K-12, E. coli C does not have any Lag phase. This reminds us, the growth pattern of Gram positive bacterium Streptococcus pneumoniae (Sp). It is well known that Professor J Lederberg has extensively used in his laboratory experiments this strain of E. coli K-12 without any adverse effect, so we think that is an avirulent strain. Question still remains that this E. coli K-12 becomes male by the presence of fertility factor F. Besides, a single chromosome of E. coli K-12, F plasmid continues as an extra chromosome, stringently controlled being a single copy of length 100 Kb [13]. In the Microbiology text book F prevails not always as extrachromosome but integrates into the chromosome, which is substantiated by Dr, William Hayes as HfrH strain [20]. What is more, an aberrant excision of F forms F-primes like F-prime lac + or F-prime trp+ and retains maleness of the host E. coli K-12. S. pneumoniae is a pathogenic strain and produces several severe diseases (Bacterial pneumonia in children and the elderly, paediatric dental caries, otitis media, meningitis). However there is a degree of difference in pathogenesis. E. coli HF4714 growth pattern is somewhat similar to that of E. coli C but not like E. coli K-12 (Figures 2 and 3).
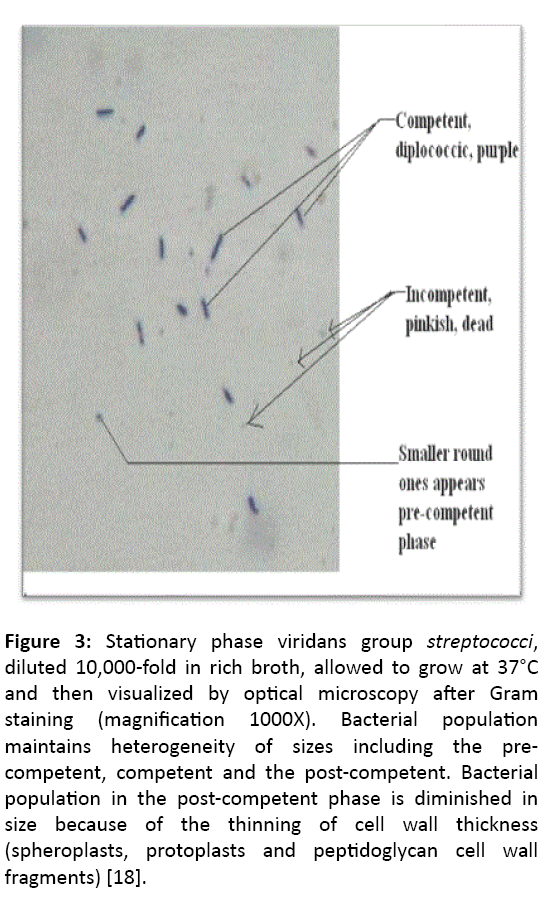
Figure 3: Stationary phase viridans group streptococci, diluted 10,000-fold in rich broth, allowed to grow at 37°C and then visualized by optical microscopy after Gram staining (magnification 1000X). Bacterial population maintains heterogeneity of sizes including the precompetent, competent and the post-competent. Bacterial population in the post-competent phase is diminished in size because of the thinning of cell wall thickness (spheroplasts, protoplasts and peptidoglycan cell wall fragments) [18].
In this work, we have compared the growth curves of Gram negative E. coli C (xylitol positive) and Dr, Lederberg’s laboratory strain of E. coli, E. coli K-12 (xylitol negative). Difference between these two strains is that E. coli C is capable of utilizing 5-carbon sugar alcohol xylitol as a carbon source and producing plaques when it absorbs the bacteriophage ɸX174. E. coli K-12, on the contrary, is apparently resistant to the xylitol and ɸX174. However, E. coli HF4714 generated by the homologous recombination between E. coli K-12 and E. coli C, is capable of metabolizing xylitol constitutively. In short, the recombinant E. coli HF4714 grows even in minimal media containing xylitol alone. E. coli HF4714 can utilize xylitol, and is resistant to bacteriophage ɸX174. Genome of bacteriophage ɸX4174 is single stranded, covalently closed circular (CCC) DNA. The phage adheres to the outer membrane of E. coli C using its spikes. Subsequently phage multiplies and releases thousands of mature particles by the lysis of E. coli C (Figures 4 and 5) [21,22].
Figure 4: Effect of 2% xylitol on viridians group Streptococcus ultrastructure as revealed by differential Gram-staining imaged with the same magnification. (a) Planktonic viridans group Streptococcus cells cultured in BHI medium and 0% xylitol anaerobically for 24 h prior to growth in xylitol. The titer has been reduced 10,000-fold to ensure that all bacteria attain log-phase growth in the presence of the xylitol [19].
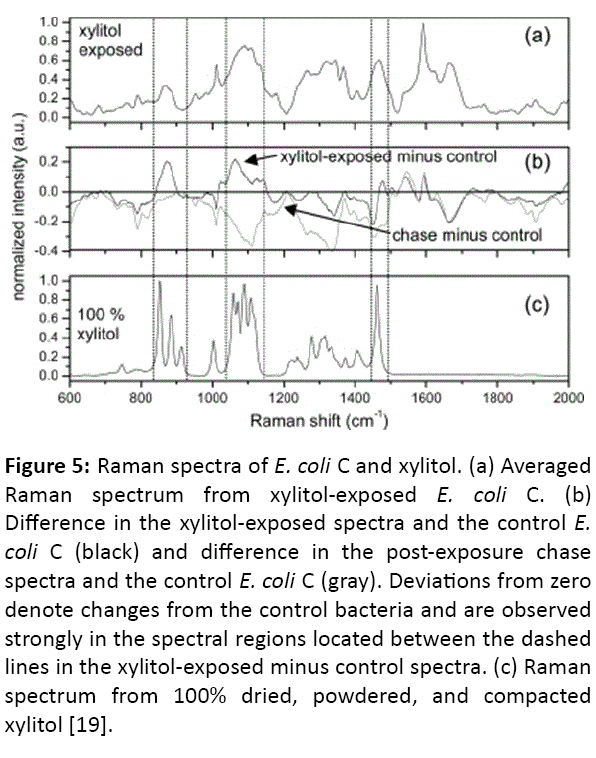
Figure 5: Raman spectra of E. coli C and xylitol. (a) Averaged Raman spectrum from xylitol-exposed E. coli C. (b) Difference in the xylitol-exposed spectra and the control E. coli C (black) and difference in the post-exposure chase spectra and the control E. coli C (gray). Deviations from zero denote changes from the control bacteria and are observed strongly in the spectral regions located between the dashed lines in the xylitol-exposed minus control spectra. (c) Raman spectrum from 100% dried, powdered, and compacted xylitol [19].
Discussion
Avirulent Gram negative E. coli C is completely lysed by the semi–temperate phage ɸX174. Based on the similarity of growth patterns, we like to postulate that the phage ɸX174 should also affect the virulent Gram positive S. pneumoniae. At the same time in our recent publications we have demonstrated that there is no such lysis. In this context we like to recall that there is a big difference between cell-wall of Gram positive S. pneumoniae and the outer membrane of Gram negative E. coli C. Below this membrane there is a single layer of peptidoglcan layer and then the inner membrane encircling the cytoplasm. There is a single chromosome of approximately 4734 Kb and at the same time the cell wall of Gram-positive S. pneumoniae consists of several peptidoglycan layers and then the inner membrane. The cell wall is formed via cross-bridges of pentapeptides [23]. Once the unfolding is initiated by their growth in presence of xylitol (2% or more), their spheroplast is formed and followed by protoplast, Cell wall is fragmented and these peptidoglycan fragments inhibit any further growth of bacteria [24].
During the reproduction of S. pneumoniaea, a cleavage is formed at the midpoint of the diplococcus and it appears as diplococcus. But it is never two bacteria, but one bacterium. Therefore the word diplococcic is misleading. However the cleavage is an index of reproduction which is very similar to humans. In 2004 review article professor Sanford A. Lacks has made a great mistake (in all his articles) by his statement that the artificial transformation of E. coli K-12 and the natural transformation of S. pneumoniae is similar [25]. Probably he has been biased by the gene cloning technique of Dr. S. N. Cohen [8-10]. Fragments of E. coli K-12 DNA (double stranded) are forcibly introduced into a recipient artificially made competent by growing in the presence of 0.01 M CaCl2 followed by a thermal shock. Major problem is the difference between such “Artificial transformation and the Natural transformation” as observed by Dr. F. Griffith in 1928. S. pneumoniae grows in three phases, pre competent, competent and post competent [2]. E. coli K-12 does not grow in chains but Pneumococci grow in chains [2]. In natural transformation, there is no role of exogenous DNA fragments. This is purely a physiological process as in humans. The oval shaped precompetent bacterium becomes competent diplococcic. In this process why the question of entry of exogenous DNA (single or double strand) arises? Such a misconception is carried over for almost 50 years starting with Avery et al. [26]. They have never made it clear that there is a difference between tiny DNA fragments (TCA insoluble) and DNA bio-macromolecule of Watson and Crick, 1953 Nobel Prize [27]. Bio-macromolecule of length 4734 Kb is the whole chromosome of E. coli K-12 and 2200 Kb approximately is the chromosome of S. pneumoniae [28-30]. Previously Cohen et al. have not made it clear that there is a big difference between a transposon and antibiotic resistant determinant. Now we have accepted that the r-det component is a collection of transposons flanked by the IS1 sequence [31]. Previously Dr. N. Kleckner has published a Review Article in Annual Review of Genetics (Vol 15, 1981) without the knowledge about the integration of F-plasmid into its host chromosome E. coli K-12 and formation of F-prime plamid (Type-1). The Type-2 uses transposons (nonhomologous and Rec-A independent) [32].
Acknowledgement
Professor W. K. Maas, New York University School of Medicine is remembered as a great teacher of Professor Sunil Palchaudhuri (SP) who has been crowned with the Hirschl Career Scientist award of 1975 in New York State. SP also acknowledges Professor M. R. J. Salton (Chairman) and Professor E. McFall of New York University School of Medicine. Dr. SP becomes Associate Professor at NYU School of Medicine within a year (1975-1976).
For the completion of this article financial assistance was provided by Atlanta Health Center for women, Kolkata, India.
References
- Griffith F (1928) The Significance of Pneumococcal types. J Hyg (Lond) 27: 113-159.
- Palchaudhuri S, Palchaudhuri A, Chatterjee B (2016) Growth curve of Streptococcus oralis. Eur J Biol Res 6: 36-41.
- Paterson GK, Blue CE, Mitchell TJ (2013) Role of two component systems in the virulence of Streptococcus pneumoniae. J Med Microbiol 55: 355-363.
- Kausc L, Mitchell M, Goodgal SH (1989) Size and physical map of the chromosome of Haemophylus influenza. J Bacteriol 171: 2472-2479.
- Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, et al. (1999) Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237: 223-234.
- Ledeberg J, Cavalli LL, Lederberg EM (1952) Sex compatibility in E. coli. Genetics 37: 720-727.
- Goulian M, Kornberg A, Sinsheimer RL (1967) Enzymatic Synthesis of DNA, XXIV. Synthesis of Infectious Phage ΦX174 DNA. Proc Natl Acad Sci USA 58: 2321-2328.
- Cohen SN, Chang ACY, Hsu L (1972) Nonchromosomal antibiotic reisitance in bacteria: Genetic transformation in E. coli by R factor DNA. Proc Natl Acad Sci USA 60: 2110-2114.
- Cohen SN, Chang ACY, Boyer HW, Helling RB (1973) Construction of biologically functional bacterial plasmids in vitro. Proc natl Acad Sci (Wash.) 70: 3240-3244.
- Cohen SN (2013) DNA cloning: A personal view after 40 years. Proc Natl Acad Sci USA 110: 15521-15529.
- Jacob F, Monod J (1965) Nobel Prize lecture.
- Kleckner N (1981) Transposable elements in prokaryotes. Ann Rev Genet 15: 341-404.
- Palchaudhuri S, Palchaudhuri A, Biswas A (2017) Mobile DNA elements in Shigella flexneri and emergence of antibiotic resistance crisis. Int J Genom Data Min 3.
- Mukhopadhyay P (1983) Novel recombination and mutation enhance the survival of E. coli K-12 under the conditions inhibitory to DNA synthesis, Ph.D. Dissertation, Wayne State University, USA.
- Gyles CL, Palchaudhuri S, Maas WK (1976) Genes for enterotoxin production and drug resistance. Science 198: 198-199.
- Kornberg HL, Lambourne LTM, Sproul AA (2000) Facilitated diffusion of fructose via the phosphoenolpyruvate/ glucose phosphotransferase system of Escherichia coli. Proc Natl Acad Sci USA 97: 1808-1812.
- Kornberg H, Lourenco C (2006) A route for fructose utilization by Escherichia coli using fucose regulon. Proc Natl Acad Sci USA 103: 19496-19499.
- Dissanayake P, Gomez-Lopez N, Czarnecki G, Palchaudhuri S (2014) Flow Cytometry Monitors Xylitol Metabolism of Streptococcus mitis. J Mol Genet Med 8: 1-5.
- Palchaudhuri S, Rehse SJ, Hamasha K, Syed T, Kurtovic E, et al. (2011) Raman spectroscopy of xylitol uptake and metabolism in Gram-positive and Gram-negative bacteria. Appl Environ Microbiol 77: 131-137.
- Hayes W (1964) The Genetics of bacteria and their viruses (2ndedn), John Wiley and Sons Inc., New York, USA.
- Palchaudhuri S, Poddar RK (1967) Influence of the viral genome on ribonucleic acid synthesis in Escherichia coli infected with bacteriophage φX174. J Mol Biol 32(3): 505-511.
- Palchaudhuri S., 1968. Ph.D. Dissertation. University of Calcutta. India.
- Ghuysen JM, Bricas E, Lache M, Leyh-Bouille M (1968) Structure of the cell walls of Micrococcus lysodeikticus. III. Isolation of a new peptide dimer, N-α-[l-alanyl-γ-(α-d-glutamylglycine)]-l-lysyl-d-alanyl-N-α-[l-alanyl-γ-(α-d-glutamylglycine)]-l-lysyl-d-alanine. Biochemistry 7: 1450-1460.
- Palchaudhuri S (2013) International EM meeting at SINP, Kolkata, India.
- Lacks SA (2004) Transformation. In: The Pneumococcus. ASM Press, Washington DC, USA.
- Avery OT, Macleod CM, McCarty M (1944) Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med 79(2): 137-158.
- Watson JD, Crick FHC (1953) Molecular structure of Nucleic acids. Nature 171: 737-738.
- Gasc AM, Kauc L, Bareile P, Sicard M, Goodgal SH (1991) Gene localization, size and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol 173: 307-312.
- Gasc AM, Giammarinaro P, Richter S, Sicard M (1996) Organization around the dnaA gene of Streptococcus pneumoniae. Microbiology 144: 433-439.
- Fuller RS, Funnel BE, Kornberg A (1984) The Dna A protein complex with the E. coli chromosomal replication region (Ori C) and other dNA sites. Cell 38: 889-890.
- Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74: 417-433.
- Palchaudhuri S, Biswas A. Mobile DNA elements: Insertion sequence and transposons. (Manuscript under preparation).
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences