ISSN : 2576-392X
Dentistry and Craniofacial Research
Association of TGFÃÆà ½Ãâò3 variants with Nonsyndromic Cleft Lip and Palate in Guatemalan Population.
Alia Said Aljabeiti, Reem Salahuddin, Miroslav Tolar and Marie M Tolarova*
Pacific Craniofacial Genetics Laboratory, University of the Pacific-Arthur A Dugoni School of Dentistry, San Francisco, CA, USA
- *Corresponding Author:
- Marie M Tolarova
Pacific Craniofacial Genetics Laboratory
University of the Pacific-Arthur A Dugoni School of Dentistry
5th Street San Francisco, CA 94122, USA
Tel: +1 (415) 786-0716
E-mail: tolarova@pacific.edu
Received Date: July 25, 2017; Accepted Date: August 14, 2017; Published Date: August 21, 2017
Citation: Aljabeiti AS, Salahuddin R, Tolar M, Tolarova MM (2017) Association of TGFß3 variants with Nonsyndromic Cleft Lip and Palate in Guatemalan Population. J Den Craniofac Res Vol.2:No.1:10. doi: 10.21767/2576-392X.100010
Abstract
Introduction: Nonsyndromic cleft lip with or without cleft palate (NCL/P) is a common orofacial anomaly with a multifactorial etiology. The genetic component of NCL/P etiology is complex with multiple genes involved. The transforming growth factor beta 3 (TFGβ3) is among the strong susceptibility genes that have been shown to be involved in morphogenesis of lip and palate.
Materials and Methods: Case-control study design was used in this study. Cases (n=237) were patients affected with NCL/P identified at the Roosevelt Hospital in Guatemala City, Guatemala. Controls (n=168) were individuals with no history of NCL/P from the same hospital. Genotypes were established by PCR and PAGE for TGFβ3 rs2300607 A/T SNP and by PCR and sequencing for TGFβ3 rs2268625 C/T SNP.
Results: For rs23000607A/T, a proportion of TT homozygotes was significantly higher (p=0.029) in cases than in controls (13.92% vs 5.36%). T allele frequency was 0.340 in cases and 0.274 in controls. Also for rs2268625 C/T, a significantly higher proportion of TT homozygotes was found among cases than among controls (p=0.041; 54.0% vs. 48.98%). Frequency of mutated T allele was 0.744 in cases and 0.694 in controls.
Conclusion: The results of our study showed for the first time the association between TGFβ3 rs2268625 C/T polymorphism and NCL/P and confirmed for Guatemalan population the association between TGFβ3 rs2300607 A/T polymorphism and NCL/P found by Ichikawa in Japanese by Ichikava et al. in 2006. Thus, our findings suggest that rs2268625 C/T and rs2300607 A/T mutations in the TGFβ3 gene are associated with NCL/P in Guatemalan population.
Keywords
Nonsyndromic cleft lip and palate TFGβ3; TGFβ3 rs2300607 A/T; TGFβ3 rs2268625; C/T Guatemala; Case-control study
Introduction
Orofacial clefts (OFC) are among the most common craniofacial malformations in humans. The prevalence rates vary between 0.5 and 3 per 1000 total births, with considerable variations between populations, genders and geographic regions [1].
In general, Asian and Amerindian populations have the highest frequencies of OFC, often at 1/500, and Africanderived populations have the lowest frequencies at 1/2500 [2]. Tolarova and Cervenka [3] analyzed the occurrence of OFC in an ethnically diverse population in California. The non-Hispanic whites had the highest birth prevalence of isolated clefts, followed by Asians and blacks who had the lowest values.
In the USA, twenty infants are born with an orofacial cleft on an average day, or 7500 every year. Children who have an OFC require several surgical procedures and complex medical treatments and, together with their families, often suffer serious psychological problems. The estimated lifetime medical cost for each child with an OFC is $100,000, amounting to $750 million for all children with OFC born each year in the USA [4].
Recently, over 30 potential candidate loci and candidate genes throughout the human genome were identified as strong susceptibility genes for OFC. The MSX1 (4p16.1), TGFα (2p13), TGFβ1 (19q13.1), TGFβ2 (1q41), TGFβ3 (14q24), RARA (17q12), MTHFR (1p36.3) genes are among the strongest candidates [5-7].
The TGFβ3 was identified as a strong candidate for clefting in humans based on a mouse model. Generally, palatogenesis in mice parallels that of humans and shows that comparable genes are involved [8]. Kaartinen et al. [9] demonstrated that mice lacking the TGFβ3 peptide exhibit cleft palate. In addition, it was shown that the exogenous TGFβ3 peptide can induce palatal fusion in chicken embryos, although the cleft palate is a normal feature in chickens [10].
In humans, association studies between the TGFβ3 gene and NCL/P have shown conflicting results. Lidral et al. [11] reported failure to observe an association of a new allelic variant of TGFβ3 with NCL/P in a case-control study of the Philippines’ population. Another study by Tanabe et al. [12] analysed DNA samples from 43 Japanese patients and compared results with those from 73 control subjects with respect to 4 candidate genes including TGFβ3. No significant differences in variants of TGFβ3 between case and control populations were observed.
On the other hand, more recent case-control association studies, family based studies, and genome scans have supported a role of TGFβ3 in cleft development. Beaty et al. [13] examined markers in five candidate genes on 269 caseparent trios ascertained through a child with non-syndromic OFC. Eighty five percent of the probands in the study were Caucasian. Markers at two of the five candidate genes (TGFβ3 and MSX1) showed consistent evidence of linkage and disequilibrium due to linkage. Similarly, Vieira et al. [14] attempted to detect transmission distortion of MSX1 and TGFβ3 in 217 South American children from their respective mothers. A joint analysis of MSX1 and TGFβ3 suggested that there might be an interaction between these two genes increasing cleft susceptibility. These results suggest that MSX1 and TGFβ3 mutations make a contribution to clefts in South American populations.
In a study of the Korean population, Kim et al. [15] reported that the G allele at the SfaN1 polymorphism of TGFβ3 is associated with an increased risk of NCL/P. The population study consisted of 28 NCL/P patients and 41 healthy controls.
In 2004, Marazita et al. [5] performed a meta-analysis of 13 genome scans of 388 extended multiplex families with NCL/P. The families came from seven diverse populations including 2,551 genotyped individuals. The meta-analysis revealed multiple genes in 6 chromosomal regions including the region containing TGFβ3 (14q24).
In the Japanese population, blood samples from twenty families with NCL/P patients have been analyzed using TGFβ3 CA repeat polymorphic marker. Based on the results of the study, the investigators concluded that either the TGFβ3 gene itself or an adjacent DNA sequence may contribute to the development of cleft lip and palate [16].
Recently, another study by Ichikawa and colleagues [17], investigated the relationship between NCL/P and seven candidate genes (TGFβ3, DLX3, PAX9, CLPTM1, TBX10, PVRL1, TBX22), in a Japanese population. The sample consisted of 112 NCL/P cases with their parents and 192 controls. Both population based case-control analysis and family based transmission disequilibrium test (TDT) were used. The results showed significant associations of SNPs in TGFβ3 and NCL/P, especially IVS+5321(rs2300607) with a p value of 0.0016. Although IVS-1572 (rs2268625) alone did not show a significant difference between cases and controls, the haplotype “A/A” for rs2300607-rs2268625 showed significant association. The author concluded that the results demonstrated positive association of TGFβ3 with NCL/P in the Japanese.
To explore a possible role of TGFβ3 polymorphism in the etiology of NCL/P in Guatemala, we investigated case and control samples from Guatemala City.
Materials and Methods
TGFβ3 gene characteristics
The TGFβ3 gene is mapped on chromosome 14, at 14q24 (Figure 1). It consists of 7 exons and 6 introns. It covers 23.91 Kb [18]. The rs2268625 C/T SNP of TGFβ3 is located in intron 1 of the gene.
Beta-type transforming growth factors form a group of polypeptides that control proliferation and differentiation of multiple cell types. TGFβ3 plays a significant role in normal fusion of palatal shelves during embryonic development (OMIM database). It is especially expressed in epithelial cells lining a medial edge of palatal shelves. It is essential for adhesion of the medial edge epithelium and elimination of the midline seam, hence providing for normal fusion of the palatal shelves. It functions similarly during fusion of facial processes forming lips. It controls angiogenesis and proliferation of mesenchymal cells [19].
Sample characteristics
Cases and controls were identified in the Roosevelt Hospital in Guatemala City, Guatemala, in 2001, 2002, 2003, and 2005. The group of cases with NCL/P consisted of 237 subjects. They were selected from a group of individuals who came to the hospital seeking treatment for various craniofacial malformations. For this study, only patients with nonsyndromic cleft lip or cleft lip and palate (NCL/P) were included in the case group.
The control sample consisted of 168 individuals with no history of birth defects who had their blood drawn at the same hospital for various follow-ups during the time of collection of cases and their specimens.
Physical and dental exams were performed for cases and controls. Personal interviews were conducted with the patients, if their age permitted, or most often with their mothers or guardians. Information on family and medical history, demographics, and digital photographs of individuals were taken. Written informed consent was obtained from individuals in both case and control groups.
Location characteristics
The Republic of Guatemala has a unique location within Central America. It is considered the geographic center of the continent. It is bordered by Mexico on the North and West, by El Salvador and Honduras on the Southeast, by Belize and the Caribbean Sea on the Northeast, and by the Pacific Ocean on the South. Guatemala occupies a total area of 108,890 sq km which is slightly smaller than Tennessee. It has a population of 12.7 million with more than half of Guatemalans being descendants of indigenous Mayan people. Spanish as well as 23 Amerindian languages are officially recognized. About 56% of the population lives below the poverty line. Coffee is the mainstay of the economy, but tourism has become the second most important source of foreign revenue in recent years. However, agriculture is still the most important part of economy. The country produces and exports sugar, bananas, fruits, vegetables, flowers, cardamom and macadamia nuts. Additionally, has developed excellent industries devoted to the assembly of clothes and electronic products, as well as the manufacture of furniture and canned goods. Also its petroleum industry is rapidly growing [20].
Collection of specimens
Intravenous blood, mainly from the cubital veins, was collected from all individuals in vacutainers with EDTA solution and was refrigerated until ready for use. Within 10 days of drawing the samples, blood was spotted on 6 cm by 10 cm sheets of filter paper using disposable transfer pipettes. A new pipette was used for each sample. Six-to-twelve blood spots were made per sheet of filter paper and two sheets were used for each sample. Blood spots were allowed to dry for 24-72 hours before being stored in individual envelopes.
DNA analysis
DNA isolation: DNA was extracted from dry blood spots on filter paper using the technique described by Polski et al. [21] for each specimen; an individual blood spot was cut from the filter paper using sterile scissors. Each spot was cut into squares approximately 1 mm × 1 mm and dropped into a sterile 1.5 mL plastic tube. One mL of sterile DNase- and RNase- free water was added to each tube and the mixtures were incubated at room temperature for 30 minutes. Then, each tube was spun at 10,000 rpm for 3 minutes and the supernatant was removed and discarded. Two-hundred μL of Chelex-100 was added and the tubes were then incubated at 56°C for 30 minutes. The tubes were then vortexed at high speed and incubated at 95°C for 8 minutes. The tubes were again vortexed, spun at 10,000 rpm for 3 minutes, and then stored at 4°C until ready for DNA amplification by Polymerase Chain Reaction (PCR).
Polymerase Chain Reaction (PCR): The fragment of DNA containing the rs2300607 A/T SNP region or the rs2268625 C/T SNP of the TGFβ3 gene was amplified by polymerase chain reaction (PCR). A reaction mixture containing 52.5% sterile H2O, 20% 5X Taq Master, 10% 10X Taq Buffer, 2% 10 mM dNTP’s, 2.5% forward primer specific for either the rs2300607 A/T SNP or the TGFβ3 rs2268625 C/T SNP, 2.5% reverse primer specific for either the rs2300607 A/T SNP or the TGFβ3 rs2268625 C/T SNP, and 0.5% Taq DNA polymerase (Qiagen) was prepared. Primer sequences are shown in Table 1 and Table 2.
| Primer Sequences for the TGFβ3 rs2300607 A/T SNP | |
|---|---|
| Forward primer | 5’-CGATGATTCATTCCACAGATT-3 |
| Reverse primer | 5’-CCTGGACAACATAGGGAGGA-3’ |
Table 1: Primer sequences used to amplify the rs2300607 A/T SNP region of the TGFβ 3 gene.
| Primer Sequences for the TGFβ3 rs2268625 C/T SNP | |
|---|---|
| Forward primer | 5’-TGAAATGGTTTAGGGAATCGT-3’ |
| Reverse primer | 5’-AAGCTCATTCCAGCCATGTC-3’ |
Table 2: The primer sequences used to amplify the rs2268625 C/T SNP region of the TGFβ3 gene.
Forty-five μL of this preparation was mixed with 5 μL of DNA isolated by the aforementioned technique in 200 μL vials. This solution underwent a specific program of temperature cycling, using a programmed Mastercycler (Eppendorf). Each cycle started at 95°C for 15 seconds, switched to 61°C for 30 seconds, and finished at 72°C for 15 seconds. This cyclic treatment was repeated 40 times.
Following PCR, a presence of the amplified DNA was confirmed using agarose gel electrophoresis. To test for contamination, a negative control sample containing 5 μL of sterile H2O in place of 5 μL of isolated DNA was mixed with the PCR mixture and run on the Mastercycler along with each batch. Following PCR, each negative was run on an agarose gel along with its corresponding batch of samples. If the agarose gel indicated the presence of DNA in the negative control, then PCR was repeated for that entire batch of samples until a true negative was achieved.
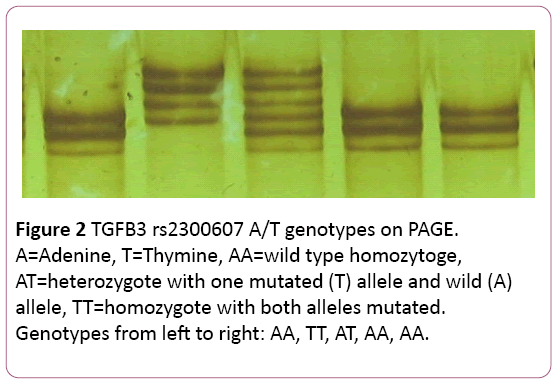
Polyacrylamide Gel Electrophoresis (PAGE): Specific patterns for AA, TT, and AT genotypes associated with rs2300607 A/T TGFβ3 gene polymorphism (Figure 2) were identified in our laboratory through Single-Stranded Conformation Polymorphism (SSCP) analysis by using a polyacrylamide gel electrophoresis [22]. Polyacrylamide gel solution was prepared by mixing 10 mL MDE (Mutation Detection Enhancement) gel monomer solution (Lonza, Inc., Rockland, ME), 2.4 mL 10X TBE buffer, 27.4 mL distilled water, 0.25 mL 10% ammonium persulfate, and 25 μL TEMED. A sample loading buffer was prepared by mixing 2.1 mL formamide, 25 μL M NaoH, 100 μL 0.5 M EDTA, 25 μL 0.02% bromophenol blue/0.02% xylene cyanol dye mixture, and 250 μL distilled water. Three μL of the loading buffer were mixed with 3 μL of each sample of amplified DNA in a 0.2 mL plastic tube. The sample/loading buffer mixture was heated to 95°C for 2-3 minutes. Four μL were then loaded into a well in the polyacrylamide gel using a fine tip pipette. A new sterile pipette tip was used for each individual sample. A negative control was run with each batch of samples to reconfirm the absence of foreign DNA contaminants in the amplified DNA. Electrophoresis was carried out at a constant power of 30 W for approximately 4 hours until the bromophenol blue dye band reached the 32 cm distance from the start wells. The gel was developed by soaking it first in 0.1% silver nitrate solution for 10 minutes, followed by soaking in a mixture of 1.5% NaOH, 0.01% sodium borohydride, and 0.15% formaldehyde for 20-30 minutes. Finally, the gel was soaked in 0.75% sodium carbonate for 3 minutes and washed in distilled water. The TGFβ3 rs2300607 A/T genotypes were determined based on a specific configuration of the stained DNA bands.
To confirm the location of bands corresponding to the TGFβ3 rs2300607 A/T polymorphism on the polyacrylamide gel, a complete sequencing of the amplified gene segment was performed for several samples. Once the genotypes were established, the samples were run on a polyacrylamide gel to display locations and patterns specific for AA homozygotes, AT heterozygotes, and TT homozygotes. These positive control samples were always run on the polyacrylamide gels along with the unknown samples to be available for comparison when genotyping was performed.
DNA purification and sequencing: We have found no specific pattern for rs2268625 C/T polymorphism using polyacrylamide gel electrophoresis (PAGE). Therefore, sequencing, preceded by purification of amplified DNA products, was conducted to determine genotypes for this single nucleotide polymorphism. QIAquick purification kit (Qiagen) was used to purify the amplified DNA products from primers, nucleotides, Taq polymerase and salts following the manufacturer’s purification protocol. The purified DNA was eluted by 40 μL of PCR water and stored at -20°C until ready for sequencing.
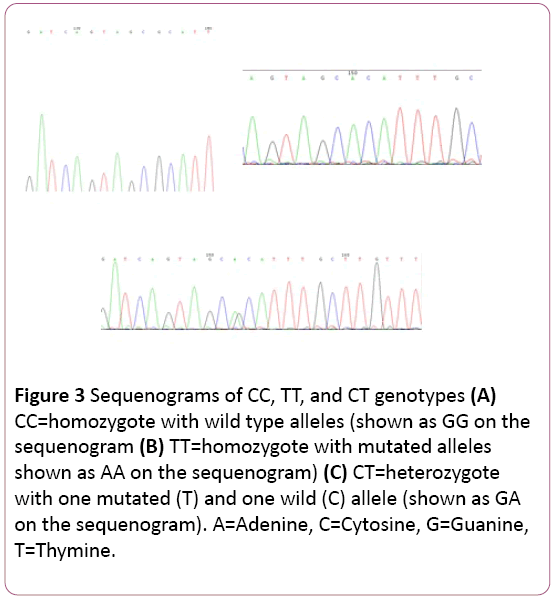
The rs2268625 C/T of TGFβ3 gene is located on the minus (complementary) strand. The sequenogram, on the other hand, is shown as a plus strand (Figures 3A-3C). Therefore, the genotypes were determined as the complementary bases of those bases presented by the sequenogram (G complements with C, A complements with T, C complements with G, and T complements with A). For example, GA on the sequenogram was indicative of CT genotype of the rs2268625 C/T polymorphism.
Figure 3 A, B, C: Sequenograms of CC, TT, and CT genotypes. CC = homozygote with wild type alleles (shown as GG on the sequenogram). TT = homozygote with mutated alleles (shown as AA on the sequenogram). CT = heterozygote with one mutated (T) and one wild (C) allele (shown as GA on the sequenogram). A= Adenine, C = Cytosine, G = Guanine, T = Thymine.
Results
The TGFβ3 rs2300607 A/T SNP (IVS+5321)
Altogether, the total of 237 cases (individuals affected with NCL/P) and 168 controls were analyzed for the TGFβ3 rs2300607 A/T polymorphism.
Distribution of TGFβ3 rs2300607 A/T genotypes in cases and controls
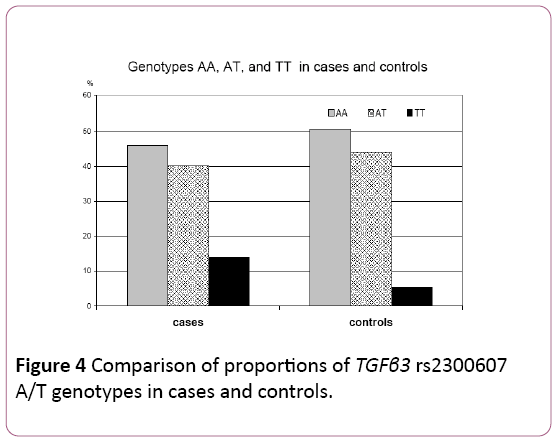
In the case sample, 109 (45.99%) individuals were homozygous for the wild type allele (AA), 95 (40.08%) individuals were heterozygous (AT) and 33 (13.93%) were homozygous for the mutated allele (TT).
In the control sample of 168 subjects, 50.59% (n=85) of control individuals were homozygotes for the wild type allele AA, 44.05% (n=74) were heterozygotes AT and only 5.36% (n=9) were homozygotes TT for the mutated allele (Table 3) (Figure 4).
| Genotypes–No (%) | ||||
|---|---|---|---|---|
| Sample type | AA | AT | TT | Total |
| Cases | 109 (45.99) | 95 (40.08) | 33 (13.93) | 237 (100.00) |
| Controls | 85 (50.59) | 74 (44.05) | 9 (5.36) | 168 (100.00) |
χ2=7.626; p=0.0206
Table 3: Distribution of TGFβ3 rs2300607 A/T genotypes in cases and controls.
The χ2 test revealed a statistically significant difference between distributions of genotypes in cases and controls (p=0.0206; χ2=0.7626).
Proportions of A and T alleles of the TGFB3 rs2300607
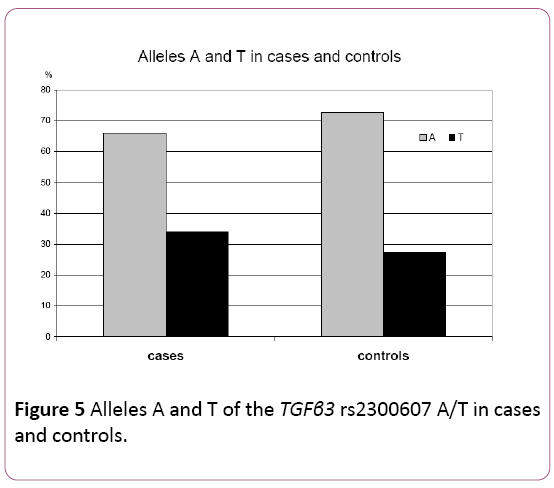
The frequency of the wild type A allele was 0.6603 in cases and 0.7262 in controls. The frequency of the T mutated allele was 0.3397 in cases and 0.2738 in the control group. Thus, there was a higher frequency of the mutated allele T in the case group as compared to the controls. The difference was on the margin of statistical significance stated for this study (p=0.0554; χ2=3.6692) (Table 4) (Figure 5).
| Alleles–No (%) | |||
|---|---|---|---|
| Sample type | A | T | Total |
| Cases | 313 (66.03) | 161 (33.97) | 474 (100.00) |
| Controls | 244 (72.62) | 92 (27.38) | 336 (100.00) |
χ 2=3.6692; p=0.0554
Table 4: A and T allele frequencies of the TGFβ3 rs2300607 in cases and controls.
The TGFβ3 rs2268625 C/T SNP (IVS-1572)
For the TGFB3 rs2268625 C/T polymorphism, 237 cases and 98 controls were analyzed.
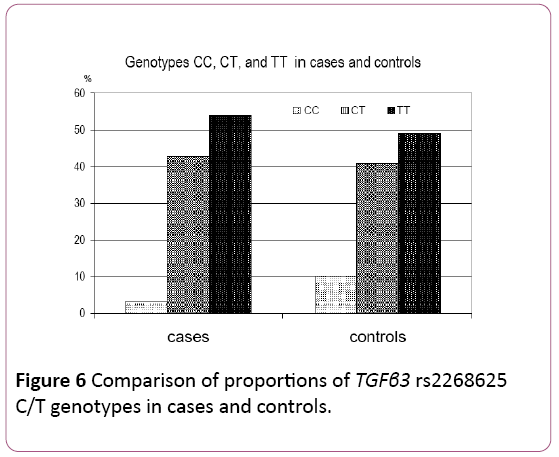
Distribution of TGFβ3 rs2268625 C/T genotypes in cases and controls: Table 5 and Figure 6 present the distribution of TGFβ3 rs2268625 C/T genotypes in cases and controls. In cases, 8 (3.38%) were homozygous for the wild type allele (CC), 101 (42.62%) were heterozygous (CT), and 128 (54.00%) were homozygous for the mutated T allele (TT). In controls, 10 (10.20%) were homozygous for the wild type allele (CC), 40 (40.80%) were heterozygous (CT), and 48 (49.00%) were homozygous for the mutated T allele (TT).
| Genotypes–No (%) | ||||
|---|---|---|---|---|
| Sample type | CC | CT | TT | Total |
| Cases | 8 (3.38) | 101 (42.62) | 128 (54.00) | 237 (100.00) |
| Controls | 10 (10.20) | 40 (40.80) | 48 (49.00) | 98 (100.00) |
χ 2=6.4038; p=0.0407
Table 5: Distribution of TGFβ3 rs2268625 C/T genotypes in cases and controls.
The distribution of TGFβ3 rs2268625 C/T genotypes significantly differs between cases and controls (p=0.041). In addition, the proportion of homozygotes for wild type allele (CC) genotype was significantly lower in cases (p=0.024) compared to controls, while proportion of mutated TT homozygotes was higher in cases.
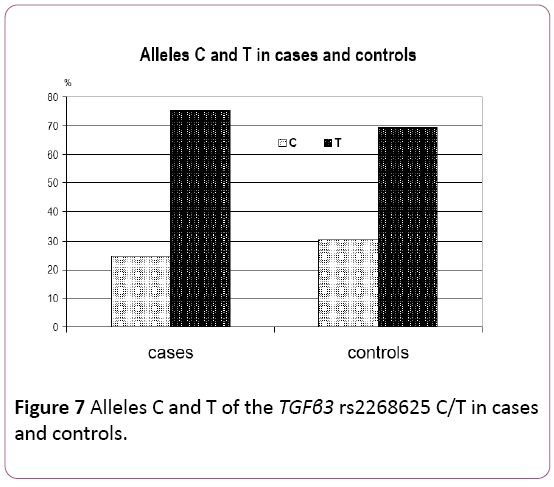
Proportions of C and T alleles of the TGFβ3 rs2268625
A comparison of the allele frequencies is shown in Table 6 and in Figure 7. The frequency of the wild type C allele was 0.2468 in the case group, compared to 0.3061 in the controls. The frequency of the T mutated allele was 0.7532 in cases, compared to 0.6939 in the control group.
| Alleles–No (%) | |||
|---|---|---|---|
| Sample type | C | T | Total |
| Cases | 117 (24.68) | 357 (75.32) | 474 (100.00) |
| Controls | 60 (30.61) | 136 (69.39) | 196 (100.00) |
χ 2=2.2115; p=0.1369
Table 6: C and T allele frequencies of the TGFβ3 rs2300607 C/T in cases and controls.
A higher frequency of the wild type C allele was observed in controls compared to cases. Although, a higher frequency of mutated allele T was observed in cases compared to controls (which may be indicating a possible role of the TGFβ3 rs2268625 C/T polymorphism in etiology of NCL/P) this difference was not statistically significant (χ²=2.2115, p=0.1369).
Combination of the TGFβ3 rs2300607 A/T and rs2268625 C/T genotypes
The distribution of combined genotypes of the TGFβ3 rs2300607 A/T and rs2268625 C/T was ascertained for 212 cases and 87 controls. The most frequent genotype combination in both cases and controls was AA/TT (46.22% in cases and 41.37% in controls) (Table 7).
| Genotypes–No (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample type | AACC | AACT | AATT | ATCT | ATTT | TTCC | TTCT | TTTT | Total |
| Cases | 0 | 2 (0.94) | 98 (46.22) | 75 (35.38) | 11 (5.19) | 13 (6.13) | 4 (1.89) | 9 (4.25) | 212 (100.00) |
| Controls | 1 (1.15) | 1 (1.15) | 36 (41.37) | 36 (41.38) | 7 (8.05) | 5 (5.75) | 1 (1.15) | 0 | 87 (100.00) |
Table 7: Distribution of combined genotypes of the TGFB3 rs2300607 A/T and rs2268625 C/T in cases and controls.
The second most frequent was AT/CT (35.38% in cases and 41.38% in controls).
We observed no combination of wild type genotypes AA/CC in cases and only one in controls. More, we did observe combination of double mutated homozygotes TT/TT only in cases (4.25%). No such genotype combination was observed in controls. There was no genotype combination AT/CC observed neither in cases nor in controls.
There were slight differences in proportions of haplotypes rs2300607 A/T and rs2268625 C/T, however none of them was found significantly different when cases and controls were compared (Table 8).
| Haplotypes–No (%) | |||||
|---|---|---|---|---|---|
| Sample type | AC | AT | TC | TT | Total |
| Cases | 79 (9.32) | 493 (58.13) | 135 (15.92) | 141 (16.63) | 848 (100.00) |
| Controls | 42 (12.07) | 196 (56.32) | 58 (16.67) | 52 (14.94) | 348 (100.00) |
Table 8: Distribution of haplotypes of the TGFB3 rs2300607 A/T and rs2268625 C/T in cases and controls.
Discussion
The identification of factors that contribute to the etiology of NCL/P is important for prevention, treatment planning, and education. With an increasing number of couples seeking genetic counseling as part of their family planning, the knowledge of how specific genes and environmental factors contribute to formation of NCL/P has gained increased importance [23]. TGFβ3 is one of several candidate genes that have been implicated in the etiology of clefts.
In this study, the distribution of TGFβ3 rs2300607 A/T genotypes and also rs2268625 C/T genotypes significantly differ between cases and controls (p=0.0206 for rs2300607 A/T and p=0.041 for rs2268625 C/T). More specifically, for rs2268625 C/T, the proportion of homozygotes for the wild type allele (CC) genotype was significantly lower in cases (p=0.024) compared to controls. In addition, a higher proportion of the mutated T allele homozygote (TT) was observed in cases compared to controls.
When allele frequencies were compared for both mutations, the higher proportion of mutated allele was found in cases compared to controls. There was a marginally statistically significant difference for TGFβ3 rs2300607 A/T and for rs2268625 C/T, even when more than three-quarters of the cases carried the mutated T allele. These results suggest that both mutations of the TGFβ3 gene are involved in the etiology of NCL/P in the population under investigation.
Ichikawa [17] investigated the possible relationship between NCL/P in a Japanese population and multiple polymorphisms of the TGFβ3 gene. The results showed significant association between NCL/P and the TGFβ3 rs2300607 A/T polymorphism. Although IVS-1572 (rs2268625) alone did not show a significant difference between cases and controls, the haplotype “A/A” for rs2300607-rs2268625 showed significant association.
The results of our study represent the first finding of an association between the TGFB3 rs2268625 C/T polymorphism and NCL/P and confirmed Ichikawa’s finding of the association between TGFβ3 rs2300607 A/T polymorphism and NCL/P.
Both studies utilized a case control design. The sample size of cases was larger in our study (n=237) compared to Ichikawa’s study (n=112), while our control sample was smaller for rs2268625 C/T (n=98) compared to 192 controls in Ichikawa’s investigation. Analysis of DNA samples from additional unaffected individuals is on-going with the aim to establish a larger control sample and to confirm our results.
Interactions between the TGFβ3 gene and other candidate genes, like RFC1, MTHFR, MSX1 and others, also need to be investigated to increase the understanding of the genetic etiological component of NCL/P. The Guatemalan population investigated in this study was studied by Costanzo et al. [24] The RFC1 A80G polymorphism was found to be associated with NCL/P in this population. Also, in the same population, MSX1 CA-repeats polymorphism was found associated with NCL/P [25]. A joint analysis of MSX1 and TGFβ3 in South American children by Vieira [14] suggested that there may be an interaction between these two genes which increases cleft susceptibility. A similar interaction between MSX1 and TGFβ3 in the Guatemalan population may need to be investigated.
The rs2300607 A/T SNP and rs2268625 C/T SNP of TGFβ3 are, interestingly, located in intron 1 of TGFβ3 gene. The mechanism of how the intronic rs2300607 A/T SNP and rs2268625 C/T SNP of TGFβ3 can contribute to clefting is still to be clarified.
It has been shown that genetic and environmental factors are ethnicity specific, and in many places in the world, location specific [23]. Thus a specific protocol for cleft prevention has to be worked out based on genetic and nutritional studies of each specific population group in order to be effective [26]. The Guatemalan population is mainly of Mayan decent as eluded earlier. Other populations with different ethnic background may have different genetic predisposition to NCL/P. Thus, future research on other populations is needed to determine the nature of the TGFβ3 gene polymorphism’s involvement in the etiology of NCL/P.
Summary and Conclusion
The etiology of NCL/P is currently considered to be the result of interactions between environmental and genetic factors. The genetic risk appears to be derived from at least several different genes. In this study, we investigated a possible role of the TGFβ3 gene in the etiology of NCL/P. Specifically, we examined the AT allele variation of TGFβ3 rs2300607A/T SNP and CT allele variation of TGFβ3 rs2268625 C/T SNP. We studied a sample of individuals from Guatemala City, Guatemala, who were affected with NCL/P and compared them to unaffected individuals from the same location.
The χ2 test revealed a statistically significant difference between distributions of TGFβ3 rs2300607 A/T SNP genotypes in cases and controls (p=0.0206). A higher proportion of mutated allele T was found in cases and this difference was on the margin of statistical significance stated for this study (p=0.0554).
A statistically significant difference in the distribution of TGFβ3 rs2268625 C/T genotypes with a higher proportion of TT homozygotes was found in cases compared to controls (p=0.041) and a lower proportion of CC homozygotes in cases compared to controls (p=0.024) A higher frequency of mutated allele T was observed in affected individuals compared to unaffected controls (this difference was not statistically significant).
The results of our study showed, for the first time, the association between the TGFβ3 rs2268625 C/T polymorphism and NCL/P and confirmed for Guatemalan population the association between TGFβ3 rs2300607 A/T polymorphism and NCL/P found by Ichikawa in Japanese [17].
Thus, our findings suggest that the rs2300607 A/T and rs2268625 C/T polymorphisms of the TGFβ3 gene are involved in the etiology of NCL/P in the Guatemalan population.
The results of this research may provide a guide for the direction of future research and genetic counseling. The potential for gene-gene and gene-environment interactions should be investigated. Further studies, preferably on larger samples, including other candidate genes and assessing haplotypes within the Guatemalan population and in other populations, are needed. Further, the identification of genes involved in causing NCL/P is the first step in formulating preventative measures that will aim to reduce the occurrence of this condition.
Acknowledgement
This research study was awarded with 2009 Harry Sicher Research Award of the American Association of Orthodontists.
The fieldwork for this study was supported and funded by Rotaplast International, Inc.
References
- Botto LD, Robert-Gnansia E, Siffel C, Harris J, Borman B, et al. (2003) International clearinghouse for birth defects monitoring systems. Annual Report 2003 with Data for 2001. Rome: The International Centre for Birth Defects 96: 774-780.
- Murray JC (2002) Gene/environment causes of cleft lip and/or palate. Clin Genet 61: 248-256.
- Tolarova M, Cervenka J (1998) Classification and birth prevalence of orofacial clefts. J Med Genet 75: 126-137.
- Waitzman NJ, Romano PS, Scheffler RM (1994) Estimates of the economic costs of birth defects. Inquiry 31: 188-205.
- Marazita ML, Murray JC, Lidral AC, Mauricio A, Cooper ME,et al. (2004) Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2p32-35. Am J Hum Genet 75: 161-173.
- Lidral AC, Moreno LM (2005) Progress toward discerning the genetics of cleft lip. CurrOpinPediatr 17: 731-739.
- Vieira AR, Avila JR, Daack-Hirsch S, Ecaterina D, Félix TM, et al. (2005) Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLos Genet 1: 64.
- Krapels IP, Vermeij-Keers C, Müller M, De Klein A, Steegers-Theunissen RP (2006) Nutrition and genes in the development of orofacial clefting. Nutr Rev 64: 280-288.
- Kaartinen V,Voncken JW,Shulter C, Warburton D, Bu D, et al. (1995) Abnormal lung development and cleft palate in mice lacking TGF-b3 indicates defects of epithelial-mesenchyme interaction. Nat Genet 11: 415-442.
- Sun D, Vanderburg CR, Odierna GS, Hay ED (1998) TGFB3 promotes transformation of chicken palate medial edge epithelium to mesenchyme in vitro. Development 125: 95-1051.
- Lidral AC, Murray JC, Buetow KH(1997) Studies of the candidate genes TGFβ2, MSX1, TGFα, and TGFβ3 in the etiology of cleft lip and palate in the Philippines. Cleft Palate Craniofac J 34: 1-6.
- Tanabe A, Taketani S, Endo-Ichikawa Y,Rikio T, Yutaka O, et al. (2009) Analysis of the candidate genes responsible for nonsyndromic cleft lip and palate in Japanese people. Clin Sci 99: 105-111.
- Beaty TH, Hetmanski JB, Zeiger JS, Fan YT, Liang KY, et al. (2002) Testing candidate genes for nonsyndromic oral clefts using a case-parent trio design. Genet Epidemiol 22: 1-11.
- Vieira AR, Romitti PA, Orioli IM, Castilla EE (2003) Complex segregation analysis of 1,792 cleft lip and palate families in South America: 1967-1997. PesquiOdontol Bras 17: 161-165.
- Kim MH, Kim HJ, Choi JY, Nahn DS (2003) Transforming growth factor beta 3 gene SfaN1 polymorphism in Korean nonsyndromic cleft lip and palate patients. J BiochemMolBiol 36: 533-553.
- Sato F, Natsume N, Machido J, Suzuki S, Kawai T (2001) Association between transforming growth factor beta 3 and cleft lip and/or palate in the Japanese population. PlastReconstrSurg 107: 1909-1910.
- Ichikawa E, Watanabe A, Nakano Y (2006) PAX9 and TGFB3 are linked to susceptibility to nonsyndromic cleft lip with or without cleft palate in the Japanese: Population-based and family-based candidate gene analyses. J Hum Genet 51: 38-46.
- https://www.ncbi.nlm.nih.gov/unigene/?term=TGF%CE%B23
- Muraoka N, Shum L, Fukumoto S, Nomura T, Ohishi M, et al.(2005) Transforming growth factor-beta3 promotes mesenchymal cell proliferation and angiogenesis mediated by the enhancement of cyclin D1, Flk-1, and CD31 gene expression during CL/Fr mouse lip fusion. Birth Defects Res A ClinMolTeratol 73: 956-965.
- https://www.state.gov/r/pa/ei/bgn/2045.html
- Polski JM, Kimzey S, Percival RW, Grosso LE (1998) Rapid and effective processing of blood specimens for diagnostic PCR using filter paper and Chelex-100. J ClinPathol: MolPathol 51: 215-217.
- Lee ST (1995) A non-radioactive method for simultaneous detection of Single-Strand Conformational Polymorphisms (SSCPs) and Heteroduplexes. Mol Cells 5: 668-672.
- Tolarová MM, Mosby T, Porter M, Olson C, Tinloy J, et al. (2007) Candidate genes and candidate nutrients in etiology of orofacial clefts. J Dent Res 86: 2991.
- Costanzo C, Tolar M, Oh HS, Grochova D, Pawar T, et al. (2004) Prevalence of RFC1 A80G mutation in cleft lip and palate. J Dent Res 83: 1308.
- Oh HS, Pawar T, Shaw A, Tolar M, Tolarová MM (2005) The role of the MSX1 gene in the etiology of non-syndromic cleft lip and palate anomalies. 7th Annual Pacific Research Day San Francisco, CA, USA.
- Tolarová MM, Poulton D, Aubert MM, Oh H, Ellerhorst T, et al. (2006) Pacific craniofacial team and cleft prevention program. J Calif Dent Assoc 34: 823-830.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences