A Pictorial Illustration of the Inhibition of Mycelial Growth and Spore Germination of Various Sorghum Fungal Pathogens by a Bacillus species
Louis K. Prom1*, Enrique G. Medrano1, Thomas Isakeit2, Roxanne Jacobsen2 and Robert Droleskey1
1USDA-ARS, Plains Area Agricultural Research Center, College Station, Texas 77845
2Department of Plant Pathology and Microbiology, Texas A & M University, College Station, TX 77843
- *Corresponding Author:
- Louis K. Prom
Department of Plant Pathology and Microbiology, USDA-ARS
Plains Area Agricultural Research Center, College Station, TX, USA
Tel: (979) 260-9393
Fax: (979) 260-9333
E-mail: louis.prom@ars.usda.gov
Received date: October 07, 2017; Accepted date: October 23, 2017; Published date: November 10, 2017
Citation: Prom L, Medrano E, Isakeit T, Jacobsen R, Droleskey R (2017) A Pictorial illustration of the inhibition of mycelial growth and spore germination of various sorghum fungal pathogens by a Bacillus species. Res J Plant Pathol 1:02.
Copyright: © 2017 Prom KL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Sorghum is one of the most indispensable cereals for food, fodder, and in brewery, especially in the drier tropics. Recently, sorghum is considered a potential source of biofuel. Globally, the productivity and profitability of sorghum is hampered by biotic stresses, causing anthracnose, grain mold, smuts, and downy mildew. In this study, a bacterium was observed growing on half-strength potato dextrose agar plate containing sorghum seeds. Using both plate and paper disc assays, activity of the determined Gram-positive Bacillus (called LP16S) was tested against four destructive sorghum pathogens Fusarium thapsinum, Colletotrichum sublineola, Curvularia lunata, and Bipolaris sp. Confirmatory in vitro analysis showed that LP16S was capable of inhibiting both mycelial growth and spore germination of these pathogens. Identification of the strain using 16S rDNA sequence analysis characterized LP16S as a putative Bacillus sp. Work is underway to determine the effectiveness of LP16S in suppressing sorghum diseases.
Keywords
Sorghum, Fusarium thapsinum, Colletotrichum sublineola, Curvularia lunata, Bipolaris sp., in vitro.
Introduction
Sorghum [Sorghum bicolor (L.) Moench] is the fifth most important crop and its uses ranged from food, feed, fodder, and recently, as a potential source of biofuel [1,2]. The crop is generally tolerant to number of adverse environmental conditions, including moisture stress and high temperature; however, its productivity and profitability across the globe in hampered by various biotic constraints, including pathogens that cause anthracnose, grain mold, smuts, and downy mildew [3-11]. Although management strategies such as the use of resistant sources has been effective in most cases in reducing the impact of these diseases, there is little information on the use of biocontrol agents as a disease management tool.
The use of biocontrol agents to combat plant diseases has long been studied, especially in vitro [12-20], but there has been little success in their application under field conditions. Ryder et al. [21] added Bacillus subtilis and B. cereus to a sodic acid soil planted with wheat in the greenhouse and reported significant reduction in the severity of take-all disease and Rhizoctonia root rot, caused by Gaeumannomyces graminis and Rhizoctonia solani, respectively. Also, the chytrid fungus (Gaertneriomyces sp.) added to soil was shown to reduce the incidence of sorghum downy mildew by 58% [22].
The use of biocontrol agents to control plant infections could be an effective strategy in situations where resistant germplasm or fungicides are not available or effective. So far, the utilization of biocontrol agents to control plant diseases has been limited due to factors such as ineffectiveness or reduced effectiveness in the field when compared to chemical fungicides, as well as commercial availability [23]. The lack of commercial availability of these agents also can be attributed to the fact that these potential biocontrol microorganisms, in particular pseudomonads and many Bacillus sp. are devoid of the capacity to produce resting spores [11] and are consequently somewhat labile.
In this communication, we show the antifungal capacity of a bacterial species that was initially observed growing on a culture plate containing sorghum seeds, producing a zone of growth inhibition of fungi. This isolate was investigated for its antagonistic potential against sorghum fungal pathogens and then identified.
Materials and Methods
Bacterial isolation
All work was conducted at the ARS-USDA-Plains Area Research Center, College Station, Texas, USA. Sorghum seeds collected from Texas AgriLife Research Farm, Burleson County, Texas, were placed on half-strength potato dextrose agar (½PDA) plates for routine analysis. After 7-10 d of incubation at 27°C, fungal growth was observed along with an apparent bacterial colony separated by a zone clear of fungi. The microorganism with the zone of inhibition was subcultured, purified, and stored in a refrigerator. Pure cultures were used for all antifungal activity testing and bacterial classification as noted below.
Screening for antifungal activity on mycelial growth
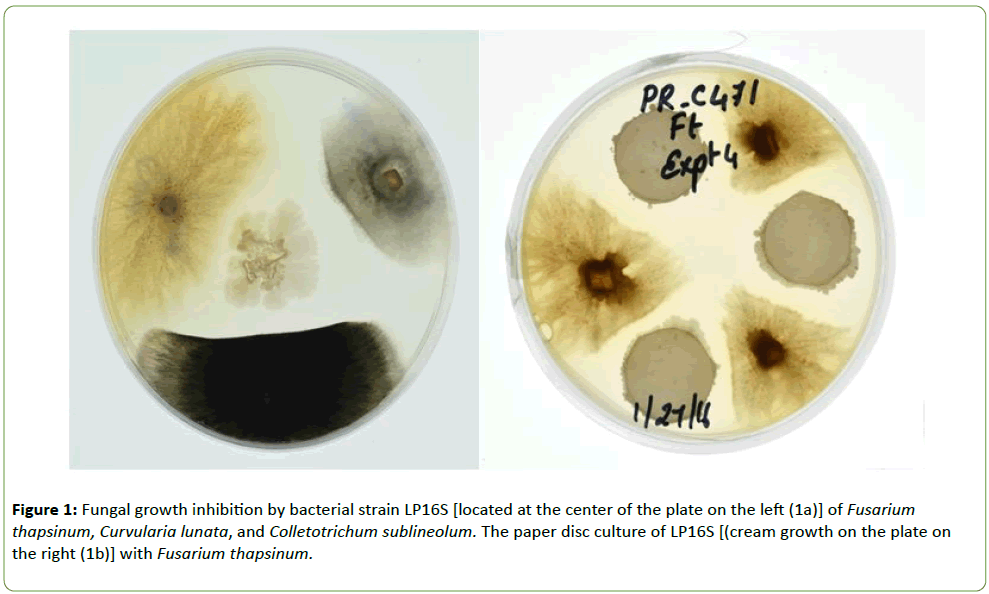
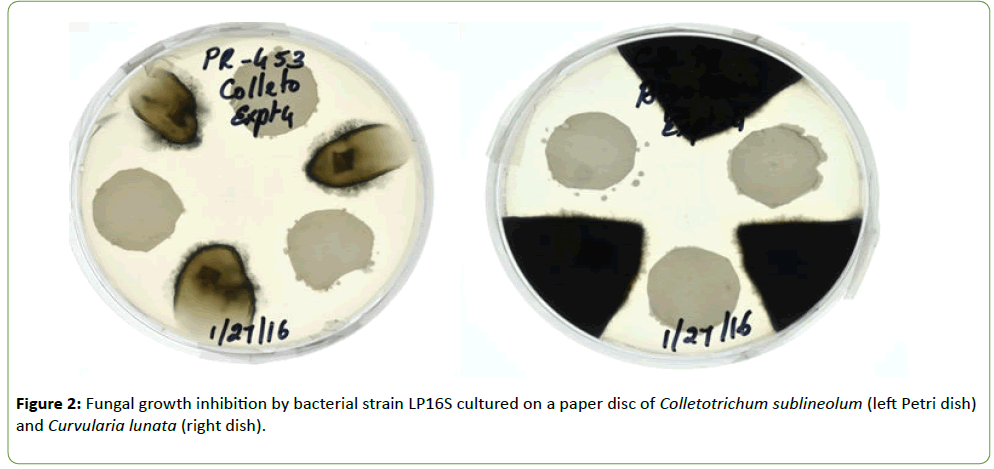
The fungal species Fusarium thapsinum, Colletotrichum sublineola, and Curvularia lunata were isolated from infected sorghum kernels and stored in a freezer at -7°C. Using the culture assay, the bacterial isolate called LP16S colony was placed in the center of a Petri dish containing ½PDA medium and agar plugs containing the different fungal species were placed on three equidistant spots (Figure 1a). In Figures 1b and 2, the paper disc assay was utilized in which three 5 mm disc dipped in Bacillus sp. spore suspension was placed in equidistant spots on Petri dish containing ½PDA and the fungal species placed between the paper disc. In both assays, the plates were incubated at 27 ± 1°C for 6 d.
Inhibition of fungal spore germination
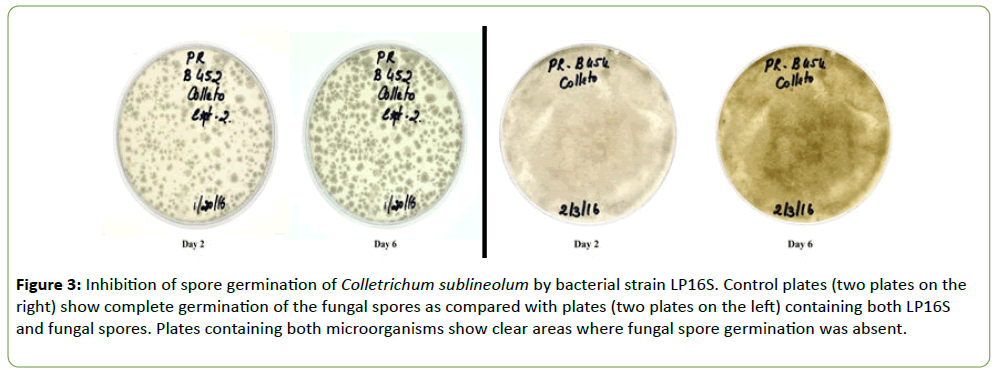
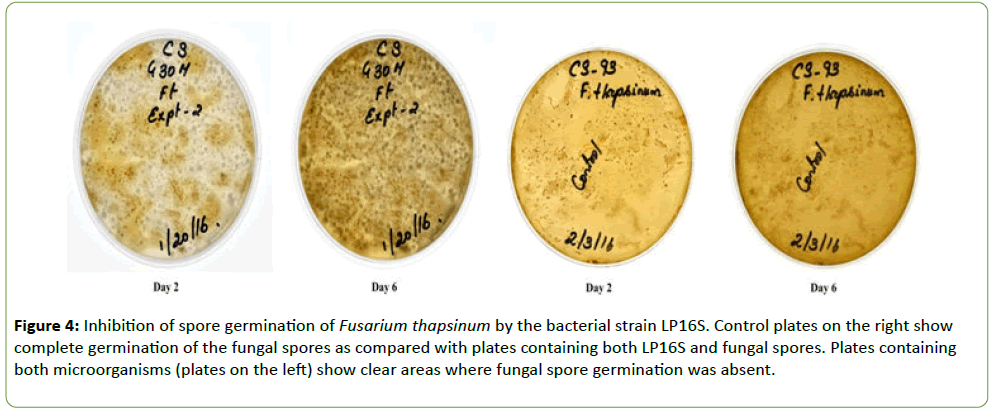
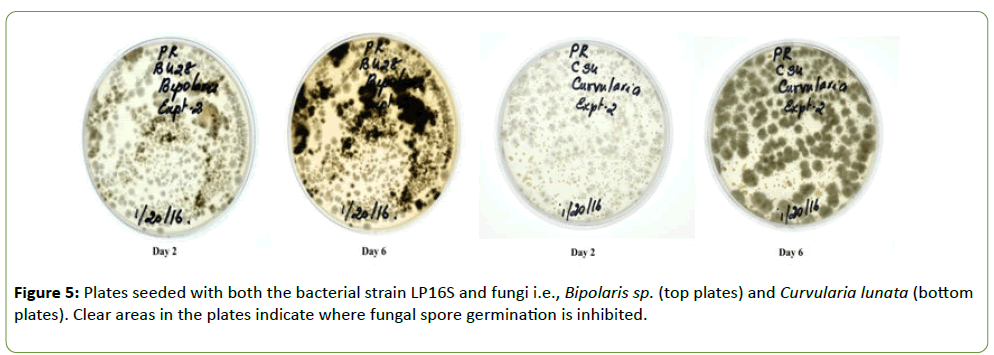
Strain LP16S was grown on Difco nutrient agar whereas the fungal isolates (F. thapsinum, C. sublineola, C. lunata, and Bioplaris sp.) were grown separately in Petri plates containing ½PDA medium. Plates were incubated at 27 ± 1°C for 6 d. LP16S spores and fungal conidia for the different isolates were harvested by flooding the plates with 10 ml sterilized water and scraping the agar surface with a rubber spatula to dislodge them. Separately, LP16S spores and the fungal conidial suspensions were filtered through two layers of sterile cheesecloth into separate beakers and diluted with sterile water to final concentrations of 1 × 106 conidia ml-1. Two drops of the LP16S spore suspension were added to four separate vials containing two drops of each fungus and mixed thoroughly. A drop of the different mixtures was spread on separate Petri dishes containing ½PDA (Figures 3-5). The control plates contained only the fungal species, C. sublineola and F. thapsinum, respectively (Figures 3 and 4). Plates were incubated at 27 ± 1°C for 6 d.
Figure 3: Inhibition of spore germination of Colletrichum sublineolum by bacterial strain LP16S. Control plates (two plates on the right) show complete germination of the fungal spores as compared with plates (two plates on the left) containing both LP16S and fungal spores. Plates containing both microorganisms show clear areas where fungal spore germination was absent.
Figure 4: Inhibition of spore germination of Fusarium thapsinum by the bacterial strain LP16S. Control plates on the right show complete germination of the fungal spores as compared with plates containing both LP16S and fungal spores. Plates containing both microorganisms (plates on the left) show clear areas where fungal spore germination was absent.
Bacterial identification
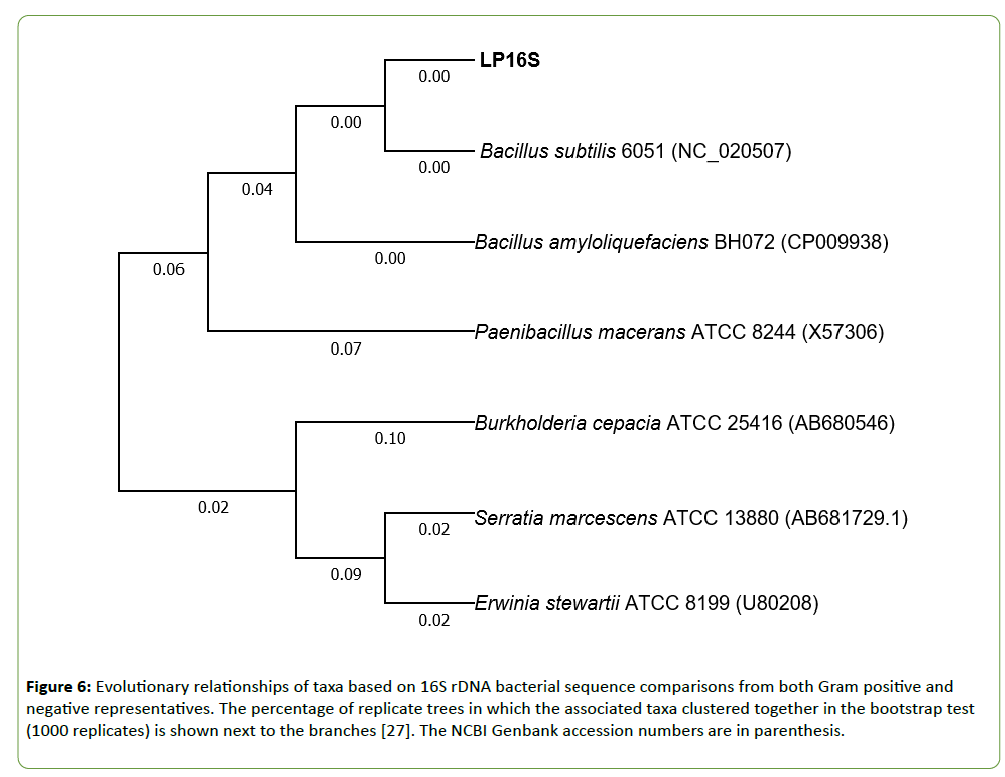
The unknown bacterium designated as LP16S was Gram stained using standard methods. The 16S rDNA sequence was generated as part of a separate whole genome sequencing study using 454 Roche Technology. Strain LP16S was characterized using the 16S rDNA bacterial sequence comparisons with both Gram positive and negative representatives. The evolutionary history generated was inferred using the Neighbor-Joining method [24]. The evolutionary distances were computed using the Jukes and Cantor [25] method and are in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA7 [26].
Results
Strain LP16S was shown to inhibit mycelial growth of F. thapsinum, C. sublineola, and C. lunata, that cause both sorghum grain mold and anthracnose. Inhibition of the mycelial growth was indicated by a clear zone between the LP16S and the fungal spp. as shown in Figures 1 and 2. Control plates containing only the fungal spp. (not shown) colonized the entire plates within the same time period. Similarly, LP16S inhibited spore germination as indicated by clear zones of the aforementioned sorghum pathogens; as well as, Bipolaris sp. (Figures 3-5). In the control plates containing only C. sublineola and F. thapsinum, the fungi grew rapidly and colonized the entire plates as shown in Figures 3 and 4.
The optimal tree with the sum of branch length=0.42228418 is shown in Figure 1. Positions containing gaps and missing data were eliminated. There were a total of 1358 positions in the final dataset. Based on the analysis, our unknown bacterium (LP16S) was characterized by 16S rDNA gene sequencing and identified as belonging to the Bacillus genus most similar to Bacillus subtilis and to a lesser degree to Bacillus amyloliquefaciens (Figure 6). The phylogenetic tree based on 16S rDNA of LP16S, two Bacillus sp., and Paenibacillus sp. showing the highest nucleotide sequence similarities is noted in Figure 6. A section of the sequencing dataset from LP16S when placed on the BLAST https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome supported similarities of the protein profile to those of B. subtilis and B. amyloliquefaciens, respectively (Figure 6).
Figure 6: Evolutionary relationships of taxa based on 16S rDNA bacterial sequence comparisons from both Gram positive and negative representatives. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches [27]. The NCBI Genbank accession numbers are in parenthesis.
Discussion
In the case of diseases that cannot be easily or economically controlled by host resistance or fungicides such as soilborne pathogens [12,13,20] or mycotoxin contamination [16], biological control may be a potential alternative.
In this study, our antagonistic bacterium (LP16S) was identified as a Bacillus sp. closely similar to B. subtilis and B. amyloliquefaciens. Antifungal activity of B. subtilis and B. amyloliquefaciens had been documented in several studies [13,15-19,21]. A number of enzymes, chemicals and antibiotics such as protease, chitinase, acetylbuanediol, iturin A, fengycin, bacillomycin, mycosubtilin, non-ribosomal lipopeptides, etc., produced by Bacillus sp. have been shown to inhibit the growth of several fungal pathogens, including C. gloeosporioides, Glomerella cingulata, Aspergillus niger, A. flavus, Rhizoctonia solani Fusarium oxysporum f. sp. cubense, and Rhizopus stolonifer [15-19]. Notably, LP16S was shown to possess similar inhibitory activity on mycelial growth and spore germination of four sorghum pathogens, F. thapsinum, C. sublineola, C. lunata, and Bipolaris sp. However, the nature of the antifungal compounds of LP16S has not yet been determined and are potentially novel. Another species possibly related to LP16S, Paenibacillus sp. obtained from the root zone of sorghum plants was shown in vitro to reduce the mycelial growth, sporangium production, zoospore germination, and germ elongation of Phytophthora parasitica [12]. In addition, the bacterium inhibited the hyphal development of F. oxysporum, F. culmorum, Aphanomyces euteiches, Chalara elegans, Pythium sp. and Rhizoctonia solani [12].
The pictorial data presented here clearly showed growth inhibition in vitro of sorghum pathogens by strain LP16S. Thus, further work towards determining the effectiveness of the putative Bacillus sp. as an antifungal biocontrol agent is ongoing. Whole genome sequencing of strain LP16S is being conducted to gain both information of probable antimicrobial products and species classification.
References
- Frederiksen RA, Odvody GN (2000) Compendium of Sorghum Diseases. 2nd ed American Phytopathological Society, St. Paul, MN. 78pp.
- Gonzalez R, Phillips R, Saloni D, Jameel H, Abt R, et al.(2011) Biomass to energy in the Southern United States: Supply chain and delivered cost. BioResources 6: 2954-2976.
- Forbes GA, Bandyopadhyay R, Garcia G (1992) A review of sorghum grain mold. In: Sorghum and Millets Diseases: A Second World Review. de Milliano WAJ, Frederiksen RA, Bengston GD, editors. International Crops Research Institute for the Semi-Arid Tropics, Patancheru, India pp 253-264.
- Frederiksen RA (2000) Head Smut In: Compendium of Sorghum Diseases. 2nd ed R. A. Frederiksen RA, Odvody GN, editors. American Phytopathological Society, St Paul, MN. pp. 18-20.
- Thakur RP, Mathur K (2000) Anthracnose In: Compendium of Sorghum Diseases. 2nd ed R A Frederiksen RA, Odvody GN, editors American Phytopathological Society, St Paul, MN. pp. 10-12.
- Thakur RP, Mathur K (2002) Downy mildews of India. Crop Protection 21: 333-345.
- King SB, Mukuru SZ (1994) An overview of sorghum, finger millet and pearl millet in eastern Africa with special attention to diseases In: Breeding for disease resistance with emphasis on durability. Danial DL, editor Wageningen Agricultural University, Wageningen, Netherlands pp 24-34.
- Bock CH, Jeger MJ, Mughoho LK, Cardwell KF, Adenle V, et al. (1998) Occurrence and distribution of Peronosclerospora sorghi [Weston and Uppal (Shaw)] in selected countries of West and Southern Africa. Crop Protection 17: 427-439.
- Gwary DM, Rabo TD, Anaso AB (2002) Assessment of leaf anthracnose caused by Colletotrichum graminicola on sorghum genotypes in the Sudan savanna of Nigeria. Agricultura Tropica et Subtropica 35: 53-58.
- Ngugi HK, King SB, Abayo GO, Reddy YVR (2002) Prevalence, incidence, and severity of sorghum diseases in western Kenya. Plant Dis 86: 65-70.
- Thakur RP, Rao V, Reddy P (2007) Downy mildew In: Screening Techniques for Sorghum Diseases. Information Bulletin no 76 Thakur RP, Reddy BVS, Mathur K, editors International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Pantacheru, India pp 31-39.
- Budi SW, van Tuinen D, Martinotti G, Gianinazz S (1999) Isolation from Sorghum bicolor mycorrhizosphere of a bacterium compatible with Arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Microbiol 65: 5148-5150.
- Weller DM (2007) Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathol 97: 250-256.
- Rosa-Magri MM, Tauk-Tornisielo SM, Ceccato-Antonini SR (2011) Bioprospection of yeasts as biocontrol agents against phytopathogenic molds. Braz Arch Biol Technol 54: 1-5.
- Islam MR, Jeong YT, Lee YS, Song CH (2012) Isolation and identification of antifungal compounds from Bacillus subtilis C9 inhibiting the growth of plant pathogenic fungi. Mycobiol 40: 59-66.
- Thakaew R, Niamsup H (2013) Inhibitory activity of Bacillus subtilis BCC 6327 metabolites against growth of aflatoxigenic fungi isolated from bird chili powder. Intern J Biosci Biochem Bioinform 3: 27-32.
- Cawoy H, Debois D, Franzil L, De Pauw E, Thonart P, et al. (2014) Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microbial Biotech 8: 281-295.
- Oyedele AO, Ogunbanwo TS (2014) Antifungal activities of Bacillus subtilis isolated from some condiments and soil. African J Microbiol Res 8: 1841-1849.
- Yuan J, Raza W, Shen Q, Huang Q (2012) Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl Environ Microbiol 78: 5942-5944.
- Shrivastava P, Kumar R, Yandigeri MS (2017) In vitro biocontrol activity of halotolerant Streptomyces aureofaciens K20: A potent antagonist against Macrophomina phaseolina (Tassi) Gold. Saudi J Biol Sci 24: 192-199.
- Ryder MH, Yan Z, Terrace TE, Rovira AD, Tang W, et al. (1999) Use of strains of Bacillus isolated in China to suppress take-all and rhizoctonia root rot, and promote seedling growth of glasshouse-grown wheat in Australian soils. Soil Biol Biochem 31: 19-29.
- Jeger MJ, Gilijamse E, Bock CH, Frinking HD (1998) The epidemiology, variability and control of the downy mildews of pearl millet and sorghum with particular reference to Africa. Plant Pathol 47: 544-569.
- Kim Y, Song,J, Lee I, Yeo W, Yun B (2013) Bacillus sp. BS061 suppresses powdery mildew and gray mold. Mycobiol 41: 108-111.
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol and Evol 4: 406-425.
- Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN, editor, Mammalian Protein Metabolism, pp. 21-132, Academic Press, New York.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol and Evol 33: 1870-1874.
- Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap Evolution. 39: 783-791.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences