HLA and KIR Gene Polymorphism in Lupus Nephritis Patients from Western India
Leenam Dedhia1,2* Vandana Pradhan1, Kanjaksha Ghosh1, Milind Nadkar3 and Sunil Parekh2
1National Institute of Immunoheamotology, 13th Floor, New Multistoreyed Building, KEM Hospital Campus, Parel, Mumbai, Maharashtra 400012, India
2Marrow Donor Registry India (MDRI), Mumbai, India, S.L. Raheja/Fortis Hospital, Old Wing, 2nd Floor, Raheja Rugnalaya Marg, Mahim (W), Mumbai, Maharashtra 400016, India
3King Edward Memorial Hospital and Seth G.S. Medical College, Acharya Donde Marg, Parel, Mumbai, Maharashtra 400012, India
- *Corresponding Author:
- Leenam Dedhia
National Institute of Immunohaematology
13th Floor, New Multistoreyed Building
KEM Hospital Campus, Parel, Mumbai, Maharashtra 400012, India
Tel: +91 22-25101305
Fax: +91 22 24138521
E-mail: leenammota@gmail.com
Received Date: January 11, 2018; Accepted Date: February 11, 2018; Published Date: February 22, 2018
Citation: Dedhia L, Pradhan V, Ghosh K, Nadkar M, Parekh S (2018) HLA and KIR Gene Polymorphism in Lupus Nephritis Patients from Western India. J Mol Sci. 2:3.
Abstract
Background: Systemic Lupus Erythematosus (SLE) is a complex multifactorial chronic autoimmune disorder characterized by humoral autoimmunity. The etiology of SLE is thought to involve an interplay of environmental, humoral, and genetic factors. Lupus nephritis (LN) is the most common clinical manifestation seen in SLE patients causing increased morbidity and mortality. There is a strong association of the human leukocyte antigen (HLA) and Killer Cell Immunoglobulin- Like Receptor (KIR) with SLE, however, the association is likely to be heterogeneous among different ethnic groups. The aim of this study was to determine the association of HLA and KIR genes in susceptibility and clinical manifestations of LN in SLE patients of western Indian population.
Methods: A total of 97 SLE patients with LN and 150 SLE patients without LN fulfilling the ACR criteria for SLE and the World Health Organisation (WHO) classification for lupus nephritis were recruited. A total of 250 age sex and ethnically matched normal healthy controls were recruited for this study.
HLA types were determined by the polymerase chain reaction on a Luminex platform with sequence-specific oligonucleotide primers (SSOP) method. Presence or absence of KIR gene polymorphism was studied using the duplex PCR sequence specific primer (SSP) technique
Results: The following HLA alleles were found to be positively associated with LN: HLA-DRB1*01 (P=0.0237), HLA DRB1*10 (P=0.0107), HLA DQB1*05(P=0.0167) and DQA1*01(P=0.0049). The HLA alleles found to be protective of LN were HLA DRB1*11 (P=0.0121) and HLA-DQB1*03 (P=.0005). Higher frequency iKIR 2DL2 was associated with LN whereas lower frequency of iKIR 2DL1, 2DL3 and aKIR2DS4 were associated with LN.
Conclusions: Our data suggest an association between HLA class II and iKIR 2DL2 with susceptibility to LN in SLE in the Indian population. HLA-DRB1*01, HLA DRB1*10, HLA DQB1*05 and DQA1*01 are strongly positively associated with SLE in western Indian SLE patients. DRB1*11 and DQB1*03 are negatively associated with SLE in western India.
Keywords
Systemic Lupus Erythematosis (SLE); Human Leukocyte Antigen (HLA); Clinical manifestations; Lupus Nephritis; HLA class II; HLA DRB1; HLA DQB1; HLA DQA1; Killer Cell Immunoglobulin-Like Receptor (KIR)
Introduction
Systemic Lupus Erythematosis (SLE) is a multifactorial autoimmune disease mostly influenced by genetic and environmental factors.
Although the exact etiology of the disease is not known, it is highly hypothesized that the accelerated rate of apoptosis and/ or impaired clearance of apoptotic bodies are the most likely causes of SLE development. SLE comprises of an inflammatory response that occurs due to the immune system attacking self-cells and tissues leading to damage. In most cases of SLE there is damage to the renal system, skin and musculoskeletal system, cardiovascular system, nervous system and respiratory system; in fact SLE can affect any tissue in the body and shows remissions and exacerbations [1].
More than 70% of patients with SLE have renal involvement at some stage of their disease. The World Health Organisation (WHO) classification for lupus nephritis has been updated for classification of LN in grades of varying severity [2]. Of the different pathological classes, diffuse proliferative glomerulonephritis (Class IV) has the worst prognosis, resulting in 11-48% of patients with end stage renal disease at 5 years.
The first genetic factor to be identified as important in the pathogenesis of SLE was the Major Histocompatibility Complex (MHC) or Human Leukocyte Antigen (HLA) complex. Defects in certain HLA genes lead to autoimmune disorders due to failure of self-tolerance. It is now widely accepted that HLA genes constitute a part of the genetic susceptibility to SLE. Human Leukocyte Antigen (HLA) and SLE has been shown to be associated since 1971 by Grumet et al. [3].
The KIRs are membrane-bound receptors found in humans and other primates. They are glycoproteins of the Immunoglobulin superfamily. In 2007, Pellett et al. [4] first reported that the frequency of KIR 2DS1 was significantly increased in SLE patients compared with controls. Since then, the association between KIR polymorphism and SLE susceptibility has been replicated in many studies, few of them being inconsistent and population specific. Thus, we performed this current study so as to evaluate the association between HLA and KIR polymorphisms and SLE.
Methods
This prospective study was conducted in 250 consecutive SLE patients from Mumbai, western India, collected over a span of three years (June 2014- June 2017). These patients were referred from Department of Medicine, KEM hospital, Mumbai, Maharashtra, India, to National Institute of Immunohaematology, (NIIH), Mumbai. All these patients were diagnosed according to the American College of Rheumatology (ACR) criteria [5]. Disease activity was assessed at the time of evaluation using Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) [6]. Pregnant and post-menopausal women, smokers, patients with diabetes and patients with significant hyperlipidemia were excluded. The patients were further divided into two groups based on the WHO criteria for the Lupus Nephritis [2]. We could clearly classify 97 patients with LN and the remaining patients were grouped as non-LN. The source of normal controls (n=250) were voluntary stem cell donors from Marrow Donor Registry India (MDRI). Blood samples (5 ml) were collected after obtaining written informed consent from all the individuals. The study protocol was approved by the Institutional Ethical Committee (IEC) of KEM hospital and NIIH.
Molecular analysis
DNA extraction was done by standard salting out technique (Bag Healthcare, Lich, Germany) followed by HLA typing using the Luminex Xmap SSOP technology (Gen-probe, CT, USA). Ambiguities of HLA genes were resolved using the Sequence Specific primer (SSP) technique (Bag Healthcare, Lich, Germany). The KIR genotyping was done using duplex SSP PCR test as previously described [7].
Statistical analysis
Continuous variables were expressed as mean+SD. Statistical analysis was carried out using fisher exact test in 2 x 2 contingency table using Graph Pad In Stat 2 software (Graph Pad Software Inc., USA). P values below 0.05 were considered statistically significant.
Results
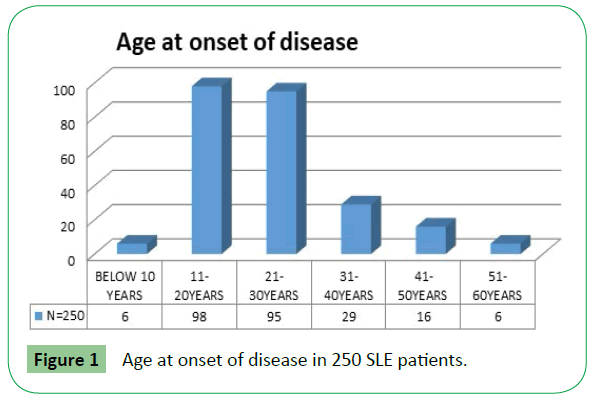
A total of 230 female patients and 20 male patients were included in the study. Their age ranged from 5-53 y (mean ± SD; 28.14 ± 9.99) in females and 7-45 y (mean ± SD; 25.75 ± 13.76) in males with about 78% patients in the age group of 11-30 years (Figure 1).
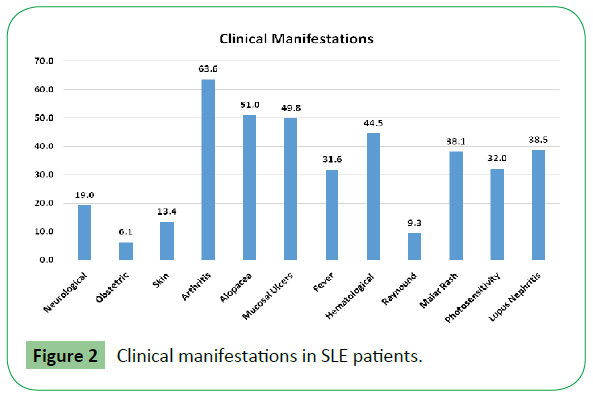
Clinical manifestation included arthritis (63.6%), alopecia (51%), mucosal ulcers (49.8%), hematological manifestations (44.5%), Lupus Nephritis (38.5%) etc. as shown in Figure 2.
We included 97 patients with LN. The allele prevalence of HLA and KIR genes were reported in percentage was compared to 250 healthy normal individuals (Table 1).
| HLA | Normal Controls | Lupus Nephritis | No Lupus Nephritis |
|---|---|---|---|
| N=250 | N=97 | N=153 | |
| % Freq | % Freq | % Freq | |
| P value v/s NC | P value v/s SG | ||
| HLA DRB1*01 | 1.4 | 5.2 | 0 |
| P=0.0237 | P=0.0061 | ||
| HLA DRB1*03 | 5.8 | 2.6 | 9.9 |
| P=0.0246 | |||
| HLA DRB1*10 | 5.1 | 12.2 | 3.3 |
| P=0.0107 | P=0.0073 | ||
| HLA DRB1*11 | 9.8 | 2.6 | 6 |
| P=0.0121 | |||
| HLA DQB1*02 | 16.4 | 11.6 | 21.4 |
| P=0.0481 | |||
| HLA DQB1*03 | 25.8 | 10.7 | 16.2 |
| P=0.0005 | |||
| HLA DQB1*05 | 21 | 32.1 | 25.3 |
| P=0.0167 | |||
| HLA DQA1*01 | 58.5 | 74.6 | 62.5 |
| P=0.0049 | P=0.0382 |
Table 1: Comparison of some polymorphic variants of HLA class II genes (% prevalence) among normal controls and SLE patients with and without Lupus nephritis (The yellow boxes highlight the KIR genes that are significant in the Normal vs. the symptomatic group and the green boxes highlight the significant difference in prevalence of KIR genes between the symptomatic and the non-symptomatic group. NC= normal control, SG= symptomatic group. The places where p value is not mentioned is not statistically significant i/e p>0.5).
We recorded a statistically significant decrease in the DRB1*11 and DQB1*03 alleles in the SLE with LN group. There was a significant increase in the DRB1*01, DRB1*10, DQB1*05 and DQA1*01 in the SLE with LN.
Within the symptomatic LN and non-symptomatic LN group the reduced frequency of DRB1*01, DRB1*10, and DQA1*01 protected patients from developing LN. Whereas increased frequencies of DRB1*03 and DQB1*02 made patients susceptible to LN (Table 2).
| KIR genes/ | Normal Controls | Lupus Nephritis | No Lupus Nephritis |
|---|---|---|---|
| frequency | N=250 | N=97 | N=153 |
| % Freq | % Freq | % Freq | |
| P value v/s NC | P value v/s SG | ||
| 2DL1 | 48 | 40 | 41 |
| P=0.0001 | |||
| 2DL2 | 27 | 36 | 37 |
| P=0.0002 | |||
| 2DL3 | 39 | 30 | 34 |
| P=0.0023 | |||
| 2DL4 | 49 | 46 | 45 |
| 2DL5 | 36 | 34 | 27 |
| P=0.0474 | |||
| 3DL1 | 43 | 37 | 39 |
| 3DL2 | 49 | 45 | 45 |
| P=0.0007 | |||
| 3DL3 | 47 | 47 | 48 |
| 2DS1 | 41 | 38 | 38 |
| 2DS2 | 33 | 32 | 34 |
| 2DS3 | 24 | 19 | 15 |
| 2DS4 | 43 | 34 | 41 |
| P=0.0002 | P=0.0092 | ||
| 2DS5 | 24 | 21 | 21 |
| 3DS1 | 27 | 28 | 30 |
Table 2: Comparison of some polymorphic variants of KIR genes (% prevalence) among normal controls and SLE patients with and without Lupus nephritis (The yellow boxes highlight the KIR genes that are significant in the Normal vs. the symptomatic group and the green boxes highlight the significant difference in prevalence of KIR genes between the symptomatic and the non-symptomatic group. NC= normal control, SG= symptomatic group. The places where p value is not mentioned is not statistically significant i/e p>0.5).
Inhibitory gene 2DL1, 2DL3, 2DL5 and 3DL 2 was significantly reduced whereas 2DL2 gene was increased in the lupus nephritis groups. In activating KIR genes, reduced frequency of 2DS4 is predisposing to lupus nephritis and higher frequency of 2DS4 is protective of lupus nephritis.
Discussion
The cause of SLE is widely unknown but several studies have presumed that it affects persons with a genetic disposition or susceptibility affected by unknown environmental triggers with defects in their immune system. The main theory that is thought to explain SLE by Lisnevskaia et al., [8] is defects in apoptotic destruction of cells.
SLE has a familial association and multiple genes such as class I, class II, and class III HLA genes appear to be responsible for a person developing lupus, this according to Martens et al., [9]. The HLA-DRB1*03 allele has been associated with SLE in Caucasian populations while the HLA-DRB1*02 allele and the HLA-DRB1*15 and HLA-DRB1*16 subtypes have been associated with SLE in black and Asian ethnic groups [10,11] Although our study did not reproduce these results owing to the different genetic and ethnic backgrounds. The prevalence of DRB1*15 is very common allele in Indian population [12,13] and this could also be a contributing factor for high prevalence of autoimmunity in Indian population as compared to the western world. HLA DRB1*04 alleles are associated with early Rheumatoid Arthritis, due to an elevated inflammatory activity and an increase of joint affections [14].
Killer cell immunoglobulin-like receptors (KIRs) belong to the immunoglobulin super family, and are expressed on both NK cells and subsets of T cells. Defects in NK function have also been demonstrated in animal models of autoimmune diseases [15,16]. Recent genetic studies have revealed that individuals carrying KIR2DL5, which is supposed to transmit inhibitory signals for NK cells, are at a decreased risk of SLE as well as an increased risk of infection [17]. In this study we reproduce these results and iKIR 2dl5 is protects patients from LN symptoms.
Conclusion
DRB1*01, DRB1*10, DQB1*05 and DQA1*01 are strongly associated in the SLE patients with LN. DRB1*11 and DQB1*03 alleles are protective in the SLE with LN group.
2DL1, 2DL3, 3DL2 iKIR genes and one activating KIR 2DS4 gene were significantly protective with LN in SLE patients
Acknowledgments
We would like to thank MDRI for the collaboration and giving access to the HLA typing instrument and kits for this study.
Conflict of interest statement: Authors have no conflict of Interest.
References
- Rahman A, Isenberg A (2008) Review Article: Systemic Lupus Erythematosus. N Engl J Med 358: 929-939.
- Weening JJ, D'Agati VD, Schwartz MM, Seshan SV, Alpers CE, et al. (2004) The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol 15: 241-250.
- Grumet A, Coukell AJ, Bodmer WF, Bodmer HO (1971) Histocompatibility (HL-A) antigens associated with systemic lupus erythematosus, a possible genetic predisposition to disease. N Engl J Med 285: pp. 193-196.
- Pellett F, Siannis F, Vukin I, Lee P, Urowitz MB, et al. (2007) KIRs and autoimmune disease: studies in systemic lupus erythematosus and scleroderma. Tissue Antigens 69: 106-108.
- Hochberg MC (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40: 1725.
- Gomez-Lozano N, Vilches C (2002) Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue antigens 59: 184-93.
- Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH (1992) Derivation of SLEDAI: a disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum 35: 630-640.
- Lisnevskaia L, Murphy G, Isenberg D (2008) Seminar: Systemic lupus erythematosus. The Lancet 384: 1878-1888.
- Martens HA, Nolte IM, van der Steege G (2009) An extensive screen of the HLA region reveals an independent association of HLA class I and class II with susceptibility for systemic lupus erythematosus. Scand J Rheumatol 38: 1-7.
- Arnett FC (1992) Genetic aspects of human lupus. Clin Immunol Immunopathol 63: 4-6.
- Doherty DG, Ireland R, Demaine AG, Wang F, Veerapan K, et al. (1992) Major histocompatibility complex genes and susceptibility to systemic lupus erythematosus in southern Chinese. Arthritis Rheum 35: 641-646.
- Dedhia L, Gadekar S, Mehta P, Parekh S (2015) HLA haplotype diversity in the South Indian population and its relevance. Indian Journal of Transplantation 9: 127-170.
- Dedhia L, Vijapukar-Valluri M, Gadekar S, Mehta P, Parekh S, et al. (2015) Distribution of HLA genes and disease predisposition in Bengali speaking people from India. Indian J Transplantation 9: 2-6.
- Seidl C, Koch U, Buhleier T, Frank R, Möller B, et al. (1997) HLA-DRB1*04 subtypes are associated with increased inflammatory activity in early rheumatoid arthritis. Br J Rheumatol 36: 941-944.
- Magilavy DB, Steinberg AD, Latta SL (1987) High hepatic natural killer cell activity in murine lupus. J Exp Med 166: 271-276.
- Nilsson N, Carlsten H (1996) Enhanced natural but diminished antibody-mediated cytotoxicity in the lungs of MRL lpr/lpr mice. Clin Exp Immunol 105: 480-485.
- Kimoto Y, Horiuchi T, Tsukamoto H, Kiyohara C, Mitoma H, et al. (2010) Association of killer cell immunoglobulin-like receptor 2DL5 with systemic lupus erythematosus and accompanying infections. Rheumatology (Oxford). 49: 1346-1353.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences