Autophagosome Maturation: Regulating Two Successive Steps
1College of Life Sciences, Key Laboratory of Agricultural and Environmental Microbiology, Ministry of Agriculture, Nanjing Agricultural University, Nanjing, China
2Department of Biochemistry and Molecular Genetics, College of Medicine, University of Illinois at Chicago, Illinois, USA
- *Corresponding Author:
- Yongheng Liang
Key Laboratory of Agriculture and Environmental Microbiology
Ministry of Agriculture, Nanjing Agricultural University, China
Tel: +553538291619
E-mail: liangyh@njau.edu.cn - Nava Segev
Department of Biochemistry and Molecular Genetics
College of Medicine, University of Illinois at Chicago
Illinois, USA
Tel: +553538291619
E-mail: nava@uic.edu
Received Date: Dec 20, 2017; Accepted Date: Dec 31, 2017; Published Date: Jan 10, 2018
Citation: Zou S, Chen Y, Zhou F, Segev N, Liang Y (2018) Autophagosome Maturation: Regulating Two Successive Steps. J Cell Dev Biol. Vol. 2 No. 1:1
Copyright: © 2018 Zou S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Plenty of information is available about molecular mechanisms that underlie the first and last steps of macroautophagy, autophagosome (AP) formation and mature AP fusion with the lysosome, respectively. In contrast, not much is currently known about factors that regulate AP maturation. Mature APs are closed doublemembrane organelles that lack phosphatidylinositol-3phosphate (PI3P) and most of the Atg proteins; removal of both is required for AP fusion. In a recent paper, we showed that the yeast Rab5-like Vps21 GTPase module is important for AP closure. In addition, we showed that this step precedes that of PI3P and Atgs removal, for which the PI3P phosphatase Ymr1 is important. Mature APs then fuse with the lysosomal membrane in a Rab7/Ypt7dependent manner. Thus, we delineate two successive steps between AP expansion and fusion: Rab5-regulated AP closure and Ymr1-regulated PI3P and Atg removal. Notably, these three players, Rab5, Ymr1 and Ypt7, are known regulators of endocytosis.
Keywords
Autophagy; Autophagosome closure; Autophagosome maturation; Rab5; Vps21; Ypt7; Ymr1; PI3P
Commentary
In macro-autophagy, the phagophore, which contains >30 Atg-related proteins, Atgs, a double membrane, and phosphatidylinositol-3-phosphate (PI3P), is formed near cargos destined for degradation [1]. The phagophore then engulfs the cargo, which can range in size, from small protein complexes to organelles, through the process of AP expansion. The cuplike double-membrane open AP has to mature before it can fuse with the vacuolar membrane to deliver the cargo for degradation [2]. AP maturation includes AP closure and release of most of the Atgs and PI3P, which were required for its formation [3,4]. Molecular mechanisms of AP formation and fusion are known. Even though AP maturation has to occur before APs can fuse with the lysosome, not much is known about the regulation of AP maturation.
Ypt/RabGTPases are known regulators of intracellular trafficking pathways including autophagy [5,6]. These GTPases function in the context of modules that include upstream activators termed guanine exchange factors (GEFs) and downstream effectors. For example, the Rab7/Ypt7 GTPase module regulates fusion of both endosome and mature autophagosome with lysosome (vacuole in yeast) in the endocytic and autophagic pathways, respectively [7]. In ypt7Δmutant cells, the closed and mature APs that disperse in the cytoplasm, lack most Atgs and PI3P. PI3P, which is required for AP formation, can be removed by the PI3P phosphatase Ymr1. Specifically, mutant cells depleted of Ymr1 accumulate closed APs decorated with PI3P and Atgs [3]. This indicates that AP closure is not sufficient for AP fusion and later maturation steps are required. Importantly, it is unknown which proteins are required for AP closure after their expansion to the normal AP size. It is expected that AP closure happens upstream of the steps regulated by Ymr1 and Rab7/ Ypt7 and that inhibition of AP closure would yield unclosed APs decorated with Atgs and PI3P.
The known function of the yeast Rab5 GTPase Vps21 is in endocytosis. We have previously shown that a Vps21/Rab5 module, which constitutes of the Vps9 GEF, the GTPase Vps21, and two of its known effectors: Vps8 and Pep12, is required for autophagy. Fluorescence microscopy and transmission electron microscopy clearly showed that under starvation, cells depleted for members of the Vps21 GTPase module accumulate AP clusters next to the vacuole [7]. However, the autophagic step defective in these mutant cells was unknown. In our recent paper, we used a protease protection assay in combination with immunoblot analysis for two cargo proteins, GFP-Atg8 and prApe1, to determine whether APs that accumulate in mutant cells are open or closed. We showed that APs that accumulate in mutants depleted of any member of the Rab5 GTPase module are unclosed. In addition, we demonstrated that AP clusters that accumulate in vps21Δmutant cells are decorated with Atgs (e.g., Atg2, 5, 18, 11, 17) and PI3P. In contrast, APs that accumulate in ymr1Δ mutant cells are closed and decorated with Atgs and PI3P, whereas APs that accumulate in ypt7Δ mutant cells are closed and lack both PI3P and Atgs [8].
The finding that APs accumulating in vps21Δ, ymr1Δ and ypt7Δ, are different, enabled us to perform a double-mutant epistasis analysis. This was important for two reasons: First, the autophagy phenotypes of vps21Δ and ymr1Δ are not complete. A possible reason for the partial autophagic phenotypes of Vps21 and Ymr1 depletion is that there are other proteins that can regulate the same steps; e.g., yeast has two additional Rab5-like GTPases and two additional PI3P phosphatases. Therefore, it was important to establish that Vps21 and Ymr1 function in the same pathway, and not in parallel pathways. Second, using this analysis, we could determine the order of events. We found that in both double mutants the vps21Δ phenotype wins. Namely, vps21Δ ymr1Δ and vps21Δ ypt7Δ double mutant cells exhibit the same phenotype as single vps21Δ mutant cells and not that of the other deletion. In both cases, the double mutant cells accumulated open APs decorated with PI3P and Atgs. These results indicate that Vps21 functions before Ymr1 and Ypt7 in the same pathway of autophagy. In agreement with this order, we showed that Ymr1 localization to APs is dependent on Vps21 [8].
The finding that APs accumulating in vps21Δ, ymr1Δ and ypt7Δ, are different, enabled us to perform a double-mutant epistasis analysis. This was important for two reasons: First, the autophagy phenotypes of vps21Δ and ymr1Δ are not complete. A possible reason for the partial autophagic phenotypes of Vps21 and Ymr1 depletion is that there are other proteins that can regulate the same steps; e.g., yeast has two additional Rab5-like GTPases and two additional PI3P phosphatases. Therefore, it was important to establish that Vps21 and Ymr1 function in the same pathway, and not in parallel pathways. Second, using this analysis, we could determine the order of events. We found that in both double mutants the vps21Δ phenotype wins. Namely, vps21Δ ymr1Δ and vps21Δ ypt7Δ double mutant cells exhibit the same phenotype as single vps21Δ mutant cells and not that of the other deletion. In both cases, the double mutant cells accumulated open APs decorated with PI3P and Atgs. These results indicate that Vps21 functions before Ymr1 and Ypt7 in the same pathway of autophagy. In agreement with this order, we showed that Ymr1 localization to APs is dependent on Vps21 [8].
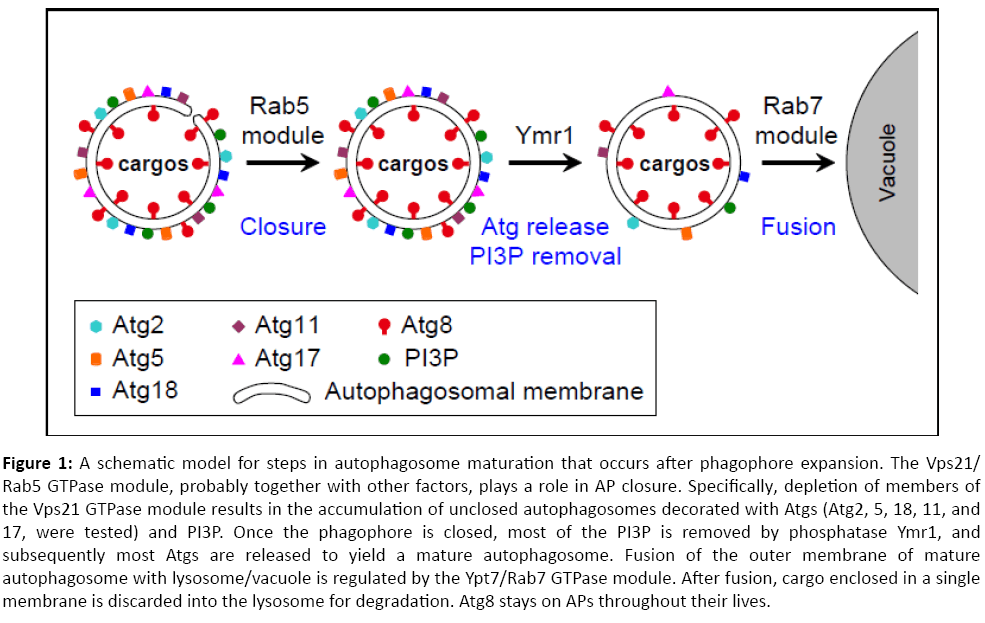
In summary, our data indicate that the Vps21/Rab5 GTPase module functions after AP expansion and upstream of AP maturation and fusion regulated by the Ymr1 phosphatase and the Ypt7/Rab7 module, respectively. We propose that the Rab5 GTPase module is important for AP closure. Once APs are closed, Ymr1 removes PI3P to facilitate the release of Atgs, which is required for generation of mature APs. Mature APs can then fuse with the lysosome in a Ypt7/Rab7-dependent way (Figure 1). Future studies should elucidate the molecular mechanism by which the Rab5 GTPase module regulates AP closure. In addition, the conservation of these steps and regulators in human cells should be explored. Importantly, the three players discussed here for their roles in AP maturation and fusion, Vps21, Ymr1 and Ypt7, have established functions in endocytosis. Therefore, our results suggest that the two pathways that lead to the lysosome, endocytosis and autophagy, merge at the Rab5-regulated step.
Figure 1: A schematic model for steps in autophagosome maturation that occurs after phagophore expansion. The Vps21/Rab5 GTPase module, probably together with other factors, plays a role in AP closure. Specifically, depletion of members of the Vps21 GTPase module results in the accumulation of unclosed autophagosomes decorated with Atgs (Atg2, 5, 18, 11, and 17, were tested) and PI3P. Once the phagophore is closed, most of the PI3P is removed by phosphatase Ymr1, and subsequently most Atgs are released to yield a mature autophagosome. Fusion of the outer membrane of mature autophagosome with lysosome/vacuole is regulated by the Ypt7/Rab7 GTPase module. After fusion, cargo enclosed in a single membrane is discarded into the lysosome for degradation. Atg8 stays on APs throughout their lives.
Acknowledgement
We are grateful to the Natural Science Foundation of China (31671479 to YL) and the National Institutes of Health (GM-45444 to NS) for funding.
References
- Weidberg H, Shvets E, Elazar Z (2011) Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80: 125-156.
- Mijaljica D, Prescott M, Devenish RJ (2012) The intriguing life of autophagosomes. Int J Mol Sci 13: 3618-3635.
- Cebollero E, Van Der Vaart A, Zhao M, Rieter E, Klionsky DJ, et al. (2012) Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Current Biology 22: 1545-53.
- Noda T, Fujita N, Yoshimori T (2009) The late stages of autophagy: how does the end begin?. Cell Death Differ 16: 984-990.
- Ao X, Zou L, Wu Y (2014) Regulation of autophagy by the Rab GTPase network. Cell Death and Differentiation 21: 348-358.
- Segev N (2001) Ypt and Rab GTPases: insight into functions through novel interactions. Current Opinion in Cell Biology 13: 500-511.
- Chen Y, Zhou F, Zou S, Yu S, Li S, et al. (2014) A Vps21 endocytic module regulates autophagy. Molecular Biology of the Cell 25: 3166-3177.
- Zhou F, Zou S, Chen Y, Lipatova Z, Sun D, et al. (2017) A Rab5 GTPase module is important for autophagosome closure. PLoS Genetics 13: e1007020.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences