ISSN : 2393-8854
Global Journal of Research and Review
Steps in Genetic Research of Complex Disease
Priya Tripathi*
Department of Internal Medicine, University of Michigan, USA
- *Corresponding Author:
- Priya Tripathi
Ph.D., Post. Doc. Fellow
Department of Internal Medicine
University of Michigan
1150 W Medical Center Drive, MSRB3
Ann Arbor, MI 48109, USA
Tel: 443-858-0480
E-mail: tripathp@med.umich.edu
Received Date: March 28, 2017; Accepted Date: March 29, 2017; Published Date: March 31, 2017
Citation: Tripathi P. Steps in Genetic Research of Complex Disease. Glob J Res Rev. 2017, 4:1.
Abstract
Many of our most common diseases are complex disorders, run in families, but they lack the simple inheritance patterns characteristics of single gene disorders. Complex diseases have low heritability compared to single gene disorders. Understanding the genetic factors underlying common disorders is a difficult but critical task because each genetic and environmental factor can contribute towards small risks. This review article will talk about steps in genetic research of complex disorders.
Keywords
Genetic research; Complex disease; Disorders
Introduction
Human diseases
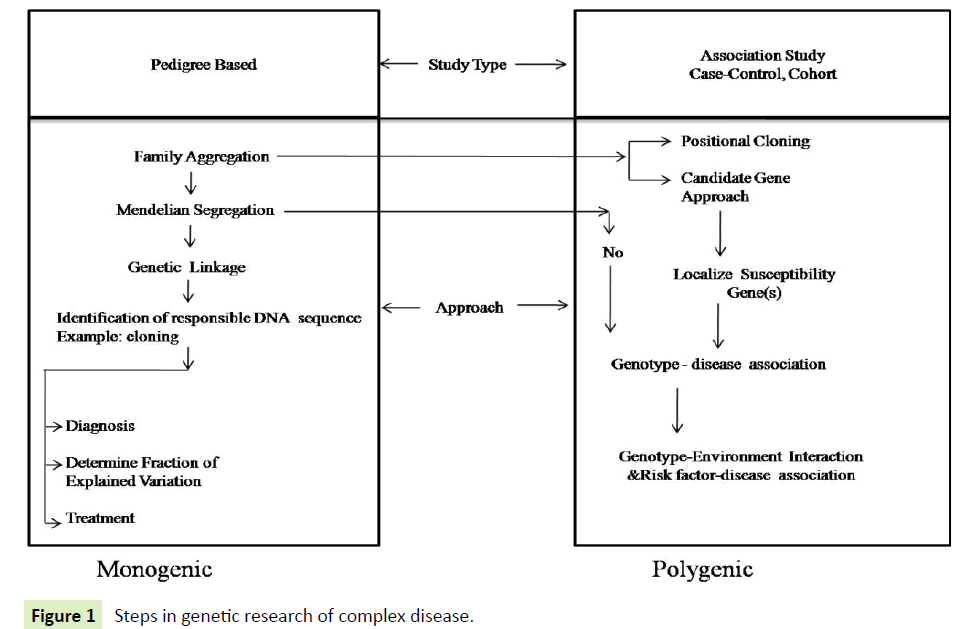
Human diseases are of two types "monogenic and polygenic". In monogenic diseases, only one gene is involved in etiology and follows the Mendelian pattern of inheritance. The phenotype of the monogenic disease may be complex (Figure 1). So many genes are measured by linkage analysis somewhere data are tranquil from affected families, the disease locus lies in the region of the genome, which is shared by all affected members of the family.
However, polygenic or common diseases are multifactorial in nature and their development results from complex interactions between numerous environmental and genetic factors (alleles of several genes). In these diseases, various components of the phenotypes result from the interaction between multiple genes and other factors. Each gene has a weak effect on phenotype, thus studies require larger sample size. Being complex in nature polygenic diseases do not obey the rule of Mendelian inheritance and pattern of inheritance is more complex than the monogenic diseases. Here, family linkage studies have been less successful.
There has been a spectacular success in identifying the genes responsible for Mendelian disorders, whereas finding the susceptibility genes involved in multifactorial diseases has been a struggle [1]. Nevertheless, how do multiple genes interact to give the final phenotype of multifactorial diseases? Genes, such as modifier genes and redundant genes that have many effects on the phenotype play an important role. Understanding the mode of action of these genes will help in determining how susceptibility genes may interact to give rise to a multifactorial phenotype [2].
Complex disorders
Our most of the common diseases are complex disorders, run in families, which lack of simple inheritance pattern characteristics or single gene disorders. These complex diseases include asthma, diabetes, stroke, other types of allergic disorders and many forms of cancers etc. Multifaceted diseases have a low heritability compared to single gene disorders. Understanding the genetic factors underlying common disorders is a difficult but critical task because each gene and environmental factor can contribute a small risk [2]. Epidemiological twin and family-based studies, as well as the animal models, provide evidence for an involvement of genetic factors in complex disorders [3,4].
Genome-wide linkage analysis and association studies are the two major ways to find out the genes responsible for complex disorders. Most of the genetic differences between individual people lie in single-nucleotide polymorphisms (SNPs). SNPs are natural variations in the four bases from which deoxyribonucleic acid (DNA) and gene are composed. It is mostly these differences that geneticist’s activity to trail chromosomes from parents to children, and in linkage analysis, to correlate the presence of certain chromosome sequences with the occurrence of disease [1]. The linkage is a phenomenon of co-segregating loci, not alleles, within families. If two markers are close, there will not be much recombination between them and they will co-segregate. This principal to definition linkage in a pedigree-based analysis. Association studies at the population level are the next step to find the mapping.
Association may result from the straight suggestion of the gene or linkage disequilibrium (LD) with the disease gene at the population level. Linkage always leads to an association but this is usually interfamilial with no association at the population level (linkage of genotype for a genetic marker to disease, may be unique to the particular family). In other words, linkage does not automatically mean a constant link with a specific allele. The allelic association may or may not be due to the linkage. Although recombination portion is what linkage studies depend on, LD is the foundation of association studies. The hypothesis is that the genetic marker studies are close enough to the actual disease gene and this will result in an allelic association at the population level.
Association studies are now a days the mainstay of genetic studies. An association study follows the candidate gene approach. The gene may be a positional or functional candidate. Positional candidate if it is located in a particular chromosome region suspected of being involved in the disease. For example; Van Eerdewegh et al. [4] identified a locus on the short arm of chromosome 20 using positional cloning and assessed 135 polymorphisms of 23 genes in this region and reported a gene encoding a disintegrin and metalloproteinase 33 (ADAM33) to be significantly associated with asthma. Genes with a known function and with the potential to cause disease is called functional candidate gene [5].
Association studies examine the frequency of specific DNA variants (alleles) in groups of unrelated individuals with the disease and unaffected controls. Association studies use case-control, cohort or family-based designs to demonstrate association in the population between particular allele and disease [6]. Limitation of an association study is that pathophysiological and biological role for studied SNPs remains unknown; investigators must identify candidate genes of interest based on its complete biological knowledge. However, advances in technology and our understanding of human genetic variations are allowing broader and more systematic surveys of possible genetic contributors to disease [6].
Conclusion
Genetic findings will central to a better taxonomy of complex diseases and the novel therapies will result from genetic findings. Collectively, an understanding important mechanistic link connecting environmental and gene factors that cause the development of these complex disorders will help in designing of personalized diagnostic tools.
References
- Todd JA (2001) Human genetics: Tackling common disease. Nature 411: 537-539.
- Burghes AH, Vaessin HE, de La Chapelle A (2001) Genetics: The land between Mendelian and multifactorial inheritance. Science 293: 2213-2214.
- Yoshinaka T, Nishii K, Yamada K, Sawada H, Nishiwaki E, et al. (2002) Identification and characterization of novel mouse and human ADAM33s with potential metalloprotease activity. Gene 282: 227-36.
- Eerdewegh PV, Little RD, Dupuis J, Del Mastro RG, Falls K, et al. (2002) Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 418: 426-430.
- Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S (2005) A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several auto immunities. Nat Genet 37: 478-485.
- Rosand J, Altshuler D (2003) Human genome sequence variation and the search for genes influencing stroke. Stroke 34: 2512-2516.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences