Molecular Identification, Prevalence and Antimicrobial Susceptibility Profile of Cronobacter spp. Cultivated on a Chromogenic Medium in Libya
Aboubaker M Garbaj1, Said K Abolghait2, Aml F Lawila1, Salah M Azwai3, Hesham T Naas1, Ashraf A Moawad4, Fatim T Gammoudi3, Ilaria Barbieri5, Salem Abureema1 and Ibrahim Eldaghayes3*
1Department of Food Hygiene and Control, University of Tripoli, Libya
2Department of Food Hygiene and Control, Suez Canal University, Egypt
3Department of Microbiology and Parasitology, University of Tripoli, Libya
4Department of Food Hygiene and Control, Cairo University, Egypt
5Experimental Zooprophylactic, Institute of Lombardy and Emilia Romagna, Brescia, Italy
- *Corresponding Author:
- Ibrahim Eldaghayes
Faculty of Veterinary Medicine
Department of Microbiology and Parasitology
University of Tripoli, P.O. Box 13662
Tripoli, Libya
Tel: +218 21 4628422
E-mail: ibrahim.eldaghayes@vetmed.edu.ly
Received Date: 13 November 2017; Accepted Date: 30 November 2017; Published Date: 07 December 2017
Citation: Garbaj AM, Abolghait SK, Lawila AF, Azwai SM, Naas HT, et al. (2017) Molecular Identification, Prevalence and Antimicrobial Susceptibility Profile of Cronobacter spp. Cultivated on a Chromogenic Medium in Libya. J Mol Microbiol. Vol. 1 No. 1: 8.
Copyright: © 2017 Garbaj AM, et al. This is an open-access article distributed under the terms of the creative Commons attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Cronobacter sakazakii is associated with illness in infants from contaminated powdered infant formula (PIF) and it is frequently recovered from PIF factory environment. Limited information is available on contamination of other food such as dairy and meat products in Libya. Methods and findings: A total of 261 samples of milk, dairy products and coarse ground meat products were collected from different localities in Libya. Samples were examined for Cronobacter spp. with an adapted ISO /DTS 22964 cultural protocol using HiChrome™ Enterobacter sakazakii modified agar coupled with 16S rDNA partial sequencing to identify the organism. The identified isolates were biochemically characterized and tested for their ability to produce yellow pigment. Out of the 261 analyzed samples, only two beef burgers, one fermented milk “Laban”, one she-camel’s milk, two raw cow’s milk, two cereal baby food, one Maassora cheese and one ready to feed baby milk were contaminated with Cronobacter spp. at a total rate of 3.8%. Accuracy of HiChrome Ent. sakazakii modified agar reach 100% as all of blue-green presumptive colonies were confirmed Cronobacter spp. while other colorless, greenish or with blue center colonies which competed growth with Cronobacter spp. were predominantly Escherichia coli followed by Klebsiella spp. and to less extent Pseudomonas luteola, Citrobacter freundii and Acinetobacter baumanii. Moreover, the isolated strains of Cronobacter were resistant to Amoxicillin, Erythromycin, Vancomycin and Streptomycin, and sensitive to Doxycycline, Enrofloxacin and Gentamycin. Conclusion: This study documents for the first time the occurrence of Cronobacter spp. in beef burger, raw cow’s milk, fermented milk “Laban”, she-camel’s milk, Maassora cheese, cereal baby food and ready to feed baby milk sold in Libya, by using conventional methods, biochemical tests and molecular techniques.
Keywords
Cronobacter spp; 16S rDNA sequence; Dairy products; Meat products; She-camel’s milk
Introduction
Cronobacter species belongs to the Enterobacteriaceae family. It is closely related to the genera Enterobacter cloacae and Citrobacter freundii [1]. It is an ubiquitous gram-negative bacteria, non-spore-forming, rod-shaped, oxidase, lactose and sorbitol negative, facultative anaerobes, motile by peritrichous flagella [2]. Recently, this pathogen has become more diverse that comprises ten species: Cronobacter sakazakii, C. malonaticus, C. turicensis, C. universalis, C. muytjensii, C. dublinensis, C. condiment, C. pulveris, C. helveticus and C. zurichensis [3].
As a hallmark distinguishing Cronobacter spp. from the rest of Enterobacteriaceae members, C. sakazakii is positive for - glucosidase enzyme, which considered the base for development of numerous chromogenic media proposed for sensitive, specific and accurate detection of this bacterium [4]. Chromogenic medium (Druggan-Forsythe-Iversen agar, DFI) is described for the selective detection of C. sakazakii. This medium is based on the -glucosidase reaction which is detected using 5-bromo-4-chloro-3-indolyl-alpha, Dglucopyranoside (X alpha Glc). C. sakazakii hydrolyses this substrate to an indigo pigment, producing blue-green colonies on this medium [5]. Commercially available biochemical test panels, such as the API 20E and ID 32E, are not sufficient to identify Cronobacter isolates at the species level and reliance on these methods will result in false positive and false negative identifications [6]. Moreover, the 16S rRNA gene sequencing showed that there is sequence differences between C. sakazakii and other Enterobacteriaceae within the hypervariable regions V1, V2, and V3 [7].
C. sakazakii is an emerging opportunistic, foodborne pathogen associated with severe illness and high mortality in neonates and infants [8]. It has also been reported in adults especially among the elderly and patients who are immunocompromised [9]. C. sakazakii has been recovered from a wide range of foods and environments; also it was isolated from human clinical samples [10].
Concerns rose due to an increasing number of cases of neonatal meningitis related to the consumption of infant formula contaminated with C. sakazakii [11]. Moreover, the International Commission for Microbiological Specifications for Foods ranked the organism as ‘severe hazard for restricted populations, life threatening or substantial chronic sequelae or long duration [12,13].
Cronobacter species are widely distributed in nature occurring in fresh water, soil and sewage, plants, vegetables, animal and human feces. Among various food samples, C. sakazakii has been found in milk and dairy products [10]. Furthermore, C. sakazakii is an important competitor to E. coli O157:H7 and Salmonella on their selective media and has recently been isolated from coarse ground beef products at contamination rates of 15% [14].
Different protein products have been prepared from milk for use in meat product formulations, these dairy protein ingredients are used to improve moisture retention, fatbinding, and textural characteristics of cooked meats. Both caseins and whey proteins have been used in comminuted and emulsified meat products, such as frankfurters and bologna, and in coarse ground meat products, such as fresh sausage, meat patties, and meatballs [15].
In Libya, consumption of raw milk and dairy products is common especially by elderly people. Cow’s milk, she-camel’s milk and locally made dairy products, such as Maassora, Ricotta and fermented milk are manufactured at small scale dairy parlors, where hygienic measures are not applied, moreover, traditionally, she-camel’s milk is consumed raw, neither pasteurized nor boiled. Studies regarding the isolation of C. sakazakii from powdered infant formula (PIF) and food products are scarce in Libya [16], which makes such a study of value not only locally but also worldwide, as infant food can be risk factor for infant illness. Therefore, the aim of this study was to investigate the prevalence of C. sakazakii in PIF, dairy and meat products in Libya with a special reference to its antibiotic resistant profile.
Materials and Methods
Collection of samples
A total of 261 samples of milk, PIF, dairy and ground meat products were randomly collected from different geographic localities in Libya (Table 1).
Table 1 Type and number of the examined samples contaminated with confirmed Cronobacter spp. (blue green colonies on HiChrome Ent. sakazakii modified agar).
| Type of Sample | Total Number of Samples | Positive Samples | |
|---|---|---|---|
| Chromogenic Agar | 16S rRNA | ||
| Raw cow’s milk | 46 | 2 (4.3%) | C. pulveris |
| Raw she camel’s milk | 5 | 1(20%) | C. sakazakii |
| Raw fermented milk (Laban) | 28 | 1(3.5%) | C. sakazakii |
| UHT milk | 8 | 0 | - |
| Yoghurt | 5 | 0 | - |
| Maassora cheese | 21 | 1(4.7%) | C. pulveris |
| Ricotta cheese | 13 | 0 | - |
| Imported soft cheese | 6 | 0 | - |
| Butter | 4 | 0 | - |
| Ice cream | 6 | 0 | - |
| Full cream milk powder | 10 | 0 | - |
| Skimmed milk powder | 6 | 0 | - |
| Ground beef | 11 | 0 | - |
| Beef burger | 12 | 2 (16.6%) | C. sakazakii |
| Powdered infant formula | 36 | 0 | - |
| Growing up formula | 18 | 0 | - |
| Ready to feed baby milk | 10 | 1(10%) | C. pulveris |
| Cereal baby food | 16 | 2 (12.5%) | C. sakazakii |
| Total | 261 | 10 (3.8%) | - |
Preparation of samples for cultivation of C. sakazakii
The culturing technique of Cronobacter spp. was performed according to the reference method ISO/TS 22964:2006 [17] for the detection of C. sakazakii in milk powder and PIF. Briefly, 25 g/mL from each sample was aseptically transferred into a sterile polyethylene stomacher bag and blended with 225 mL of Buffer peptone water (Park Scientific, M 0063, Northampton Limited, UK), homogenized in a stomacher (Stomacher 400, Seaward Medicals, UK) at 230 rpm for 1 min, then incubated at 37°C for 18 ± 2 h. Ten mL of the preenrichment was cultured by inoculation into 90 mL of Enterobacteriaceae enrichment broth (EEB, Lot: 082610202 – Liofilchem- Italy) then incubated at 37°C for 24 h. Only 0.1 mL of the selective enriched broth was streaked onto a chromogenic media (HiChrome Ent. sakazakii modified agar, HiMedia, M1641, India); the inoculated plates were incubated at 44°C for 24 h. The presumptive colonies (blue green) were picked up for further investigation. HiChrome™ Enterobacter sakazakii modified agar contains a chromogenic substrate (5- Bromo-4-chloro-3-indolyl α-D-glucopyranoside) which is cleaved specifically by C. sakazakii resulting in the formation of blue green colonies [5]. Other organisms, which do not cleave this substrate, produce colorless, green or colorless with blue center colonies. Further, a presumptive C. sakazakii isolates were streaked onto Tryptone soya agar (TSA, Park, # M 266, U.K) incubated at 25°C for at least 72h to detect yellow pigmented colonies produced by C. sakazakii [18].
Biochemical characterization
Hi25™ Enterobacteriaceae identification kit (KB003 Hi 25TM Enterobacteriaceae identification kit, HiMedia) was used according to the manufacturer’s instruction.
Identification of C. sakazakii by PCR and partial sequencing of 16S rDNA
Suspected colonies cultivated on HiChrome Ent. sakazakii modified agar were picked up and purified several times. The procedure of DNA extraction of Cronobacter isolates was carried out as described before [19]. Partial 16S rDNA was amplified using the universal oligonucleotides primers; Forward: S-D-Bact-0341-b-S-17 and Reverse: S-D-Bact-0785-a- A-21 [20]. The amplified 16S rDNA PCR fragment (464 bp) was excised from the gel, and the DNA was extracted from the gel using GF-1 AmbiClean kit (Cat. # GF-GC-100, Vivantis, Malaysia). The purified 16S rDNA amplicons were then sequenced in IZSLER - Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna, Brescia, Italy.
BLAST search was carried out for the obtained consensus sequences by both NCBI (https://www.ncbi.nlm.nih.gov/pubmed) and 16S bacterial cultures Blast Server for the identification of prokaryotes (https://bioinfo.unice.fr/blast/).
Antimicrobial susceptibility profile
PCR confirmed Cronobacter isolates were tested against nine antibiotics by applying the disc-diffusion method as described in the Clinical and Laboratory Standards Institute (CLSI) [21]. These antibiotics included Gentamycin (Mast discs 10 μg), Streptomycin (Oxoid 10 μg), Amoxicillin (Arcomex 25 μg), Colistin (Mast discs 10 μg), Oxytetracycline (Oxiod 30 μg), Doxycycline (Mast discs 30 μg), Vancomycin (Oxoid 30 μg), Enrofloxacin (Arcomex 5 μg) and Erythromycin (Mast discs 10 μg).
Results
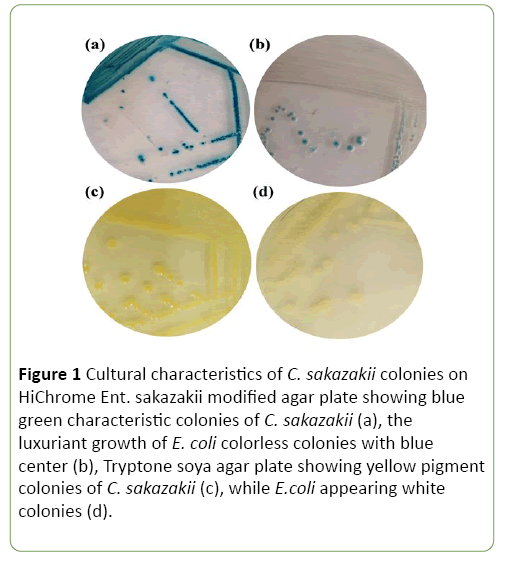
A total of 261 isolates were successfully cultivated on HiChrome Ent. sakazakii modified agar. Ten of them showed the characteristic blue-green colonies, while the rest of isolates ranged from colorless to greenish, and colorless with blue center colonies (Figure 1).
Figure 1: Cultural characteristics of C. sakazakii colonies on HiChrome Ent. sakazakii modified agar plate showing blue green characteristic colonies of C. sakazakii (a), the luxuriant growth of E. coli colorless colonies with blue center (b), Tryptone soya agar plate showing yellow pigment colonies of C. sakazakii (c), while E.coli appearing white colonies (d).
Out of 261 samples of milk, baby food, dairy and ground meat products only 10 (3.8%) yielded characteristic blue-green colonies on HiChrome Ent. sakazakii modified agar suggested to be C. sakazakii (Figure 1). Three samples out of 80 baby food samples were contaminated with C. sakazakii at a rate of (3.7%). Among the examined dairy products samples, C. sakazakii was recovered from one raw fermented milk (Laban) (3.5%), one she-camel’s milk (20%), two cow’s milk (4.3%) and one Maassora cheese (4.7%) respectively. Only two out of twelve examined beef burger samples (16.6%) were found to contain C. sakazakii (Table 1).
Moreover, only three characteristic isolates were identically C. sakazakii and produced yellow pigment colonies (Figure 1). All positive samples subjected to biochemical tests (Hi25TM identification kit, HiMedia) were positive for xylose and cellobiose but gave negative results to nitrate, indole and phenylalanine deamination tests (Table 2).
Table 2 Biochemical characterization of Cronobacter species.
| Biochemical Test | Isolate Code | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 10305.2 | 10322.1 | 10324.1 | 10429 | 10456 | 10460.6 | 10492 | 2204 | 6208 | |
| OPNG | + | + | + | + | + | + | + | + | + |
| Lysine utilization | - | - | - | - | - | - | - | - | - |
| Ornithine utilization | + | - | - | - | - | + | - | + | - |
| Urease | - | - | - | - | - | - | - | - | - |
| Phenylalanine deamination | - | - | - | - | - | - | - | - | - |
| Nitrate reduction | - | - | - | - | - | - | - | - | - |
| H2S production | - | - | - | - | - | - | - | - | - |
| Citrate utilization | + | - | - | - | - | + | - | + | + |
| Vogesproskauers | + | - | - | - | - | + | - | + | - |
| Methyl red | - | + | + | + | + | - | + | - | + |
| Indole | - | - | - | - | - | - | - | - | - |
| Malonate utilization | - | + | + | + | + | + | + | + | + |
| Esculin hydrolysis | - | + | + | + | + | + | + | + | + |
| Arabinose | - | + | + | + | + | - | + | - | + |
| Xylose | + | + | + | + | + | + | + | + | + |
| Adonitol | - | - | - | - | - | - | - | - | - |
| Rhamnose | + | - | - | - | - | + | - | + | - |
| Cellobiose | + | + | + | + | + | + | + | + | + |
| Melibiose | + | + | + | + | + | + | + | - | - |
| Saccharose | - | - | - | - | - | + | - | - | + |
| Raffinose | - | - | - | - | - | + | - | + | - |
| Trehalose | - | + | + | + | + | - | + | - | + |
| Glucose | - | + | + | + | + | + | + | - | + |
| Lactose | + | + | - | - | - | + | + | - | + |
| Oxidase | - | - | - | - | - | - | - | - | - |
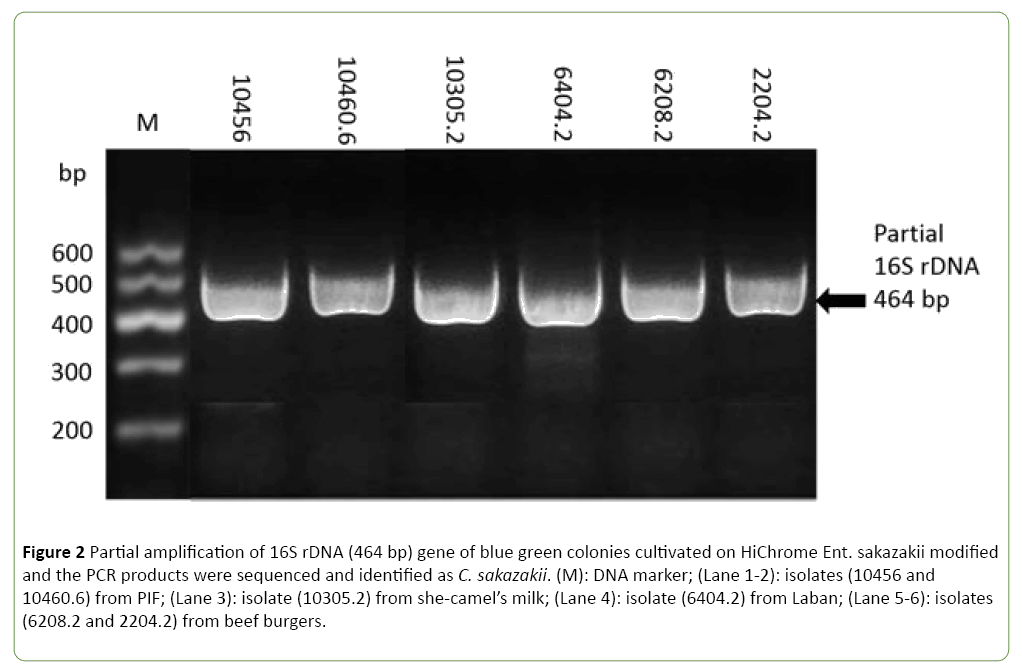
All purified isolates were subjected to PCR and partial sequencing of 16S rDNA to identify the presumptive bacteria and to ensure the selectivity of the used medium. Interestingly, this assay confirmed all above isolates as Cronobacter spp. with 99% nucleotide identity. In particular six isolates were identified as C. sakazakii, while the remaining four were C. pulveris. She-camel’s milk and beef burger could represent an important vehicles for transmission of C. sakazakii at rate of contamination of 20% and 16.6%, respectively (Table 1 and Figure 2).
Figure 2: Partial amplification of 16S rDNA (464 bp) gene of blue green colonies cultivated on HiChrome Ent. sakazakii modified and the PCR products were sequenced and identified as C. sakazakii. (M): DNA marker; (Lane 1-2): isolates (10456 and 10460.6) from PIF; (Lane 3): isolate (10305.2) from she-camel’s milk; (Lane 4): isolate (6404.2) from Laban; (Lane 5-6): isolates (6208.2 and 2204.2) from beef burgers.
Many bacterial species appeared colorless to colorless with blue center and greenish colonies were competing growth with C. sakazakii on HiChrome Ent. sakazakii modified agar. However, bacterial identification on basis of 16S rRNA Blast server for the identification of prokaryotes showed that Escherichia coli was predominant followed by Klebsiella spp. and to less extent Pseudomonas luteola, Citrobacter freundii and Acinetobacter baumanii (Table 3).
Table 3 Bacterial species that compete growth with C. sakazakii on HiChrome Ent. sakazakii modified agar as identified on basis of 16S rRNA Blast server for the identification of prokaryotes.
| Bacterial Species | Number of Isolates | Colony Color |
|---|---|---|
| Escherichia coli | 76 | Colorless with blue center |
| Klebsiella spp. | 12 | Slightly violet |
| Cronobater pulveris | 4 | Slightly violet |
| Pseudomona luteola | 3 | Brownish |
| Citrobacter freundii | 3 | Slightly violet |
| Acinetobacter baumanii | 1 | Slightly violet |
Antimicrobial susceptibility profile of C. sakazakii isolates was evaluated by using 9 antibiotics against Cronobacter spp. Results showed in Table 4 revealed that the isolates were sensitive to Doxycycline, Enrofloxacin, Gentamycin, Colistin and Oxytetracycline with the exception of two isolates that were resistant to Colistin and Oxytetracycline, respectively. In contrast, all isolates were resistant to Amoxicillin, Erythromycin, Vancomycin and Streptomycin (Table 4).
Table 4 Antimicrobial susceptibility profile of the isolated C. sakazakii.
| Isolate Code | Type of Antibiotics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMX | CO | DOX | ENR | E | G | OXT | VA | St | |
| 10305.2 | R | S | S | S | R | S | S | R | R |
| 10322.1 | R | S | S | S | R | S | R | R | R |
| 10324.1 | R | S | S | S | R | S | S | R | R |
| 10429 | R | S | S | S | R | S | S | R | R |
| 10429 | R | S | S | S | R | S | S | R | R |
| 10460.6 | R | S | S | S | R | S | S | R | R |
| 10492 | R | R | S | S | R | S | S | R | R |
| 2204.2 | R | S | S | S | R | S | S | R | R |
| 6208.2 | R | S | S | S | R | S | S | R | R |
| 6404.2 | R | S | S | S | R | S | S | R | R |
AMX: Amoxicillin, CO:Colistin, DOX:Doxycylin, ENR:Enerofloxacin, E: Erythromycin, G; Gentamycin, OXT:Oxytetracycline, VA:Vancomycin, St: Streptomycin; S: Sensitive, R: Resistance.
Discussion
Cronobacter strains were recovered from a widespread range of foods including dried foods, meat, milk, cheese, vegetables, water, tea, rice, spices and herbs, various food production environments and household [22-27]. This wide spectrum of Cronobacter-contaminated foods covers both raw and processed foods and the type of food processing is not limited to dry products [23].
Our study revealed that beef burger samples represent a considerable source of C. sakazakii, this may be attributed to the spices added to enhance flavor or due to milk powder which is used as functional dairy protein ingredients in this product. This is consistent with recent survey study showing that 16% (8/50) and 15% (6/40) of ground beef and beef burger samples were contaminated with C. sakazakii [14].
The contamination of dairy products may occur during handling, processing, inadequate refrigeration or poor personal hygiene and probable use of polluted water in cleaning of dairy utensils [28]. C. sakazakii was occasionally detected in (6.6%) of examined raw cow’ milk based only on the conventional methods and biochemical tests [28]. Other investigators found only (0.5%) of bulk tank cow’s milk samples were contaminated with C. sakazakii [29].
As shown in Table 1 and Figure 1, C. sakazakii was found in two samples (4.3%) of raw cow’s milk and one sample (20%) of raw she-camel’s milk. While the observed results in this study were slightly lower than those reported before [28] where C. sakazakii (6.6%) was isolated in raw cow’ milk based only on the conventional methods and biochemical tests. These findings were higher than that recorded by another study [29] were only (0.5%) of C. sakazakii strain was detected in bulk tank cow’s milk. Moreover, seven out of 14 samples of shecamel’s milk were contaminated by C. sakazakii which were identified on the basis of their cultural characteristics and the biochemical reactions [30]. In contrast, in other studies, C. sakazakii were not detected in cow’s milk samples [31,32].
Interestingly, this is the first report that documents the contamination of she-camel’s milk with C. sakazakii in Libya. These results supported those studies that reported C. sakazakii is a possible cause of diarrhea, wound infections, and urinary tract infections among immunocompromised people, particularly the elderly [33]. This is very important especially in developing countries such as Libya where raw she-camel’s milk is traditionally consumed without pasteurization or boiling.
Results of the present study showed that C. sakazakii was not detected in examined samples of UHT milk. The reason for this may be due to its heat treatment. However, another study reported the presence of C. sakazakii in UHT milk [28]. On the other hand, one sample out of 28 samples of fermented milk (Laban) was contaminated with C. sakazakii and none of the tested 5 samples of yoghurt contained any C. sakazakii, these results are similar with that reported in another study [34]. A possible reason for C. sakazakii not being detected in yoghurt samples could be due to its sensitivity to low pH [10]. In addition, C. sakazakii could not be detected from ice cream as previously described [9,35]. As indicated from some researchers that C. sakazakii is unable to grow in ice cream during frozen storage [10]. Although C. sakazakii had been reported in ice cream samples at range of (4% - 26.6%) [10,28].
In the current study, only one sample of tested Maassora cheese was contaminated by C. sakazakii. This finding was lower than that reported in other studies [2,9,10,28] where C. sakazakii was observed in (30%), (4%), (40%) and (23.3%) of cheese samples respectively. In contrast, other studies failed to isolate this bacterium from cheese samples [34,36].
The present investigation showed that baby food (cereal baby food and ready to feed baby milk) was contaminated with C. sakazakii at rate of (3.7%). This finding is in agreement with another study that found C. sakazakii in two cereal-based infant drinks [37]. However, no C. sakazakii was found in wheat-based infant food [30]. Moreover, C. sakazakii was not detected in milk powder samples because of its heat treatment used in the final stage which could eliminate most of pathogenic bacteria [38].
The contamination of cereal baby food and ready to feed baby milk with C. sakazakii may result from addition of some ingredients sensitive to the heat after pasteurization step such as dried fruit, also the infant rice cereal when reconstituted with water, milk or infant formula supports the growth of C. sakazakii [39].
Molecular confirmation of the recovered Cronobacter isolates was done by application of the PCR assay based on the partial amplification of 16S rDNA and using of universal oligonucleotides primers where the specific 464 bp band has been documented according with that reported before for the same genus Cronobacter using the same specific primers and 16S rDNA PCR protocol [20]. Results showed in Table 1 confirming the first report for detection of Cronobacter spp. by cultural and molecular techniques from baby food, milk and meat product samples collected from different localities in Libya. The partial sequencing of 16S rDNA confirmed that all 10 isolates were Cronobacter spp. with 99% nucleotide identity. Only 6 out of 261 (2.2%) isolates were C. sakazakii that obtained from raw she-camel’s milk, cereal baby food, beef burger and fermented milk (Laban) while the remaining 4 (1.5%) isolates were C. pulveris that were isolated from raw cow’s milk, Maassora cheese and ready to feed baby milk.
First isolation of Cronobacter pulveris was from baby food [40]. It was suggested that these isolates have a similarity with C. sakazakii depending on the chromogenic media, biochemical tests and comparative assay between 16S rRNA and rpoB gene sequence analysis. Later they re-classified these strains as a novel species Cronobacter pulveris.
The antibiotics susceptibility test of the Cronobacter spp. revealed that most isolates were resistant to most of the tested antibiotics. This is an alarm indicating the increasing resistance of Cronobacter spp. to common used antibiotics and raise concerns for the possible consequence of that on public health.
Conclusion
In conclusion, this work on genus Cronobacter, reveals the important consideration for the food industry since this bacterium can cause severe illness to exposed people in particular the highly vulnerable neonates, infants and the elderly. The presence of Cronobacter spp. in dairy and meat products considered as potential risk to the public health and transmission vehicle of this organism, especially in countries such as Libya. In this study, ten Cronobacter isolates were recovered from 261 samples of milk and milk products based on the conventional method, yellow pigmentation and biochemical tests. These results indicate that conventional method, yellow pigmentation and commercially biochemical test panels are not sufficiently reliable for speciation of Cronobacter isolates at the species level and reliance on these methods will result in false positive and false negative identifications. Furthermore, the application of molecular technique plays a major role in identification of bacteria; also it improves and facilitates the control of most pathogenic bacterial disease including the foodborne disease. Interestingly, this assay could differentiate between species in the same genus. Moreover, this is first document reporting the presences of C. sakazakii in she-camel’s milk. This is very important especially in developing countries such as Libya where raw she-camel’s milk is traditionally consumed as raw milk without heat treatment.
Acknowledgment
Authors are grateful to Veronica Papini, a technician in IZSLER - Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna, Brescia, Italy, who performed the sequencing of the partial 16S rDNA. Authors are grateful to all stuff of Food and Drug Control Center with special thanks to Eng. Abdalrhman Almerimey, the Director of Tripoli branch, Asma Ashour Beiass, Food and Drug Laboratory, and all Microbiology team.
Funding
This study was part of a project titled “Genetic authentication of bacterial isolates from meat and milk products in Libya and establishing the Foodborne Libyan-type Bacterial Collection (FLBC)” that was supported by a grant provided by the Authority of Natural Science Research and Technology (Libyan Authority for Research, Science and Technology).
References
- Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, et al. (2012) Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J Clin Microbiol 50: 3031-3039.
- Gökmen M, Tekinşen KK, Gürbüz Ü (2010) Presence of Enterobactersakazakii in milk powder, whey powder and white cheese Produced in Konya. Kafkas Üniversitesi Veteriner Fakültesi Dergisi 16: 163-166.
- Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P (2013) Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottianimipressuralis comb. nov. and Lelliottiaamnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibactergergoviae comb. nov. and Pluralibacterpyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakoniacowanii comb. nov., Kosakoniaradicincitans comb. nov., Kosakoniaoryzae comb. nov. and Kosakoniaarachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacterzurichensis nom. nov., Cronobacterhelveticus comb. nov. and Cronobacterpulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. SystApplMicrobiol 36: 309-319.
- Cawthorn DM, Botha S, Witthuhn RC (2008) Evaluation of different methods for the detection and identification of Enterobactersakazakii isolated from South African infant formula milks and the processing environment. Int J Food Microbiol 127: 129-138.
- Iversen C, Druggan P, Forsythe S (2004) A selective differential medium for Enterobactersakazakii, a preliminary study. Int J Food Microbiol 96: 133-139.
- Jackson EE, Forsythe SJ (2016) Comparative study of Cronobacter identification according to phenotyping methods. BMC Microbiol 16: 146.
- Hassan AA, Akineden Ö, Kress C, Estuningsih S, Schneider E, et al. (2007) Characterization of the gene encoding the 16S rRNA of Enterobactersakazakii and development of a species-specific PCR method. Int J Food Microbiol 116: 214-220.
- Amalaradjou MA, Kim KS, Venkitanarayanan K(2014) Sub-inhibitory concentrations of trans-cinnamaldehyde attenuate virulence in Cronobactersakazakii in vitro. Int J Mol Sci 15, 8639-8655.
- El-Sharoud WM, O'Brien S, Negredo C, Iversen C, Fanning S, et al. (2009) Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiol 9: 24.
- El-Gamal MS, El Dairouty RK, Okda AY, Salah SH, El-Shamy SM (2013) Incidence and Interrelation of Cronobactersakazakii and Other Foodborne Bacteria in Some Milk Products and Infant Formula Milks in Cairo and Giza Area. World Appl Sci J 26: 1129-1141.
- Muytjens HL, Zanen HC, Sonderkamp HJ, Kollee LA, Wachsmuth IK, et al. (1983) Analysis of eight cases of neonatal meningitis and sepsis due to Enterobactersakazakii. J Clin Microbiol 18: 115-120.
- Cai XQ, Yu HQ, Ruan ZX, Yang LL, Bai JS, et al. (2013) Rapid detection and simultaneous genotyping of Cronobacter spp. (formerly Enterobactersakazakii) in powdered infant formula using real-time PCR and high resolution melting (HRM) analysis. PLoS One 8: e67082.
- ICMSF (2006) Microorganisms in Foods 7: Microbiological Testing in Food Safety Management. Springer, US.
- Mohammed MA, Sallam KI, Tamura T (2015) Prevalence, identification and molecular characterization of Cronobactersakazakii isolated from retail meat products. Food Cont 53: 206-211.
- Tarté R (2009) Ingredients in meat products: Properties, functionality and applications. Springer, New York.
- Matug SM, Aidoo KE, Elgerbi AM (2015) Microbiological examination of infant food and feed formula. Emerg Life Sci Res 1: 46-51.
- International Organization for Standardization (2006) Milk and milk products. Detection of Enterobactersakazakii.ISO,Switzerland. pp: 1-13.
- Mullane NR, Murray J, Drudy D, Prentice N, Whyte P (2006) Detection of Enterobactersakazakii in dried infant milk formula by cationic-magnetic-bead capture. Appl Environ Microbiol 72: 6325-6330.
- Azwai SM, Alfallani EA, Abolghait SK, Garbaj AM, Naas HT (2016) Isolation and molecular identification of Vibrio spp. by sequencing of 16S rDNA from seafood, meat and meat products in Libya. Open Vet J 6: 36-43.
- Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, et al. (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME J 5; 1571-1579.
- David OM, Falegan CR, Oluyege OA (2013) Pattern of breastfeeding and occurrence of Cronobactersakazakii in infant formula sold in Ekiti State, Nigeria. Int J Curr Microbiol Appl Sci 2: 11.
- Edelson-Mammel SG, Porteous MK, Buchanan RL (2005) Survival of Enterobactersakazakii in a dehydrated powdered infant formula. J Food Prot 68: 1900-1902.
- Friedemann M (2007) Enterobactersakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 116: 1-10.
- Gurtler JB, Kornacki JL, Beuchat LR (2005) Enterobactersakazakii: A coliform of increased concern to infant health. Int J Food Microbiol 104: 1-34.
- Iversen C, Forsythe S (2004) Isolation of Enterobactersakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol 21: 771-777.
- Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M (2004) Occurrence of Enterobactersakazakii in food production environments and households. Lancet 363: 39-40.
- Turcovský I, Kuniková K, Drahovská H, Kaclíková E (2011) Biochemical and molecular characterization of Cronobacter spp. (formerly Enterobactersakazakii) isolated from foods. Antonie van Leeuwenhoek 99: 257-269.
- Saad NM, Wahba NM (2006) Risk Profile of Enterobactersakazakii in Raw and UHT Milk and Some Milk Products. Assuit University, Egypt. pp: 931- 947.
- Jayarao BM, Wang L (1999) A study on the prevalence of gram-negative bacteria in bulk tank milk. J Dairy Sci 82: 2620-2624.
- Kalpana (2014) Characterization and control of camel milk isolates of cronobactersakazakii. M. Sc Thesis, Deemed University.
- Hochel I, Ruzickova H, Krasny L, Demnerova K (2012) Occurrence of Cronobacter spp. in retail foods. J Appl Microbiol 112: 1257-1265.
- Baumgartner A, Niederhauser I (2010) Occurrence of Cronobacter spp. in raw milk. J VerbrLebensm 5: 253.
- Friedemann M (2009) Epidemiology of invasive neonatal Cronobacter (Enterobactersakazakii) infections. Eur J Clin Microbiol Infect Dis 28: 1297-1304.
- El-Sharoud WM, El-Din MZ, Ziada DM, Ahmed SF, Klena JD (2008) Surveillance and genotyping of Enterobactersakazakii suggest its potential transmission from milk powder into imitation recombined soft cheese. J Appl Microbiol 105: 559-566.
- Baumgartner A, Grand M, Liniger M, Iversen C (2009) Detection and frequency of Cronobacter spp. (Enterobactersakazakii) in different categories of ready-to-eat foods other than infant formula. Int J Food Microbiol 136: 189-192.
- Meshref AMS, Hassan GM (2009) Bacteriological status of some soft cheeses sold in Beni-Suef city. Assiut Vet Med J 55: 112-123.
- O'Brien S, Healy B, Negredo C, Anderson W, Fanning S, et al. (2009) Prevalence of Cronobacter species (Enterobactersakazakii) in follow-on infant formulae and infant drinks. Lett Appl Microbiol 48: 536-541.
- Putthana V, Marounek M, Brenova N, Mrazek J, Lukesova D (2012) Isolation and characterization of Cronobacter spp. from environmental and food resources. Agriculturatropica et subtropica 45: 5-11.
- Huang Y, Pang Y, Wang H, Tang Z, Zhou Y, et al. (2015) Occurrence and Characterization of Cronobacter spp. in Dehydrated Rice Powder from Chinese Supermarket. PLoS One 10: e0131053.
- Stephan R, Van Trappen S, Cleenwerck I, Iversen C, Joosten H, et al. (2008) Enterobacterpulveris sp. nov., isolated from fruit powder, infant formula and an infant formula production environment. Int J Sys Evol Microbiol 58: 237-241.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences