Bacterial Degradation of Petroleum and Petroleum Products
Mihaela Marilena Stancu*
Institute of Biology Bucharest of Romanian Academy, Bucharest, Romania
- *Corresponding Author:
- Mihaela Marilena Stancu
Institute of Biology Bucharest of Romanian Academy
296 Splaiul Independentei, Bucharest 060031
P.O. Box 56-53, Romania
Tel: +40212219202
Fax: 40212219071
E-mail: mihaela.stancu@ibiol.ro
Received Date: 16 March 2018; Accepted Date: 19 March 2018; Published Date: 26 March 2018
Citation: Stancu MM (2018) Bacterial Degradation of Petroleum and Petroleum Products. J Mol Microbiol. Vol. 2 No. 1: 2.
Copyright: © 2018 Stancu MM. This is an open-access article distributed under the terms of the creative Commons attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abbreviations
MBC: Mixed Bacterial Consortia; PBC: Pure Bacterial Culture; GT: Growth Test; MF Metabolic Fingerprinting; LM: Light Microscopy; TEM: Transmission Electron Microscopy; SEM: Scanning Electron Microscopy; ARDRA: Amplified Ribosomal DNA Restriction Analysis; RAPD: Random Amplified Polymorphic DNA; REP-PCR: Repetitive Extragenic Palindromic Polymerase Chain Reaction; T-RFLP: Terminal Restriction Fragment Length Polymorphism; PFGE: Pulsed Field Gel Electrophoresis; Q-PCR: Quantitative PCR or Real-Time PCR; RT-Q-PCR: Reverse Transcription Q-PCR; SDS-PAGE: Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis; HPTLC: High Performance Thin Layer Chromatography; DGGE/TGGE: Denaturing/Temperature Gradient Gel Electrophoresis; LHPCR: Length Heterogeneity PCR; RISA: Ribosomal Intergenic Spacer Analysis; SSCP: Single Strand Conformation Polymorphism; FGA: Functional Gene Arrays; FISH: Fluorescence In Situ Hybridization; FAME: Fatty Acid Methyl Esters; DNA: Deoxyribonucleic Acid; RNA: Ribonucleic Acid
Editorial
Petroleum is the major source of energy used in several industries and in our daily life. Subsequently, the release of petroleum hydrocarbons into the environment due to accidental spills or human activities, such as oil exploitation, transportation, and storage are the main causes of water and soil pollution [1,2]. Petroleum is a complex mixture of different hydrocarbons including alkanes, aromatics, asphaltenes, resins, and majority of these compounds are very toxic and carcinogenic for living organisms [1,3-5]. Therefore, the cleaning up of petroleum hydrocarbons in the water and soil environment is a real world problem. Some of the microorganisms, primarily bacteria which naturally exist in water and soil, are capable to degrade toxic petroleum hydrocarbons under different environmental conditions [1,5]. Furthermore, bacteria exposed to such toxic compounds become adapted, exhibiting higher biodegradation rates, as compared with bacteria from unpolluted sites [2,3,6]. Several adaptive mechanisms (e.g., modifications of the membrane or changes in the overall energy status, modifications of cell morphology and cell surface properties, active excretion, induction of anabolic pathways) were described for bacteria able to survive in the presence of toxic petroleum hydrocarbons [7-9].
Bioremediation represents the use of bacteria to degrade toxic petroleum hydrocarbons into non-toxic end products, such as carbon dioxide, water and other inorganic compounds, and these processes are an environmentally friendly, efficient, economic and versatile alternative to physicochemical treatment of water and soil polluted with petroleum hydrocarbons [3-5,10-12]. Generally, bioremediation technologies are classified as in situ or ex situ. In situ bioremediation (e.g., natural attenuation, bioaugmentation, biostimulation) involves treating the pollutants at the site, while ex situ (e.g., landfarming, composting, biopiles, bioreactors) involves the removal and transporting of the pollutants to another site for treatment [13-15].
It is well known, that the behavior of petroleum hydrocarbons in the environment is dependent by the nature and concentration of the hydrocarbons present and by the interaction between environmental and biological factors [6]. Hydrocarbons differ in their susceptibility to bacterial attack, and biodegradability of the hydrocarbons generally decreases in the following order: linear alkanes, branched alkanes, monoaromatics, cyclic alkanes, polyaromatic hydrocarbons, and asphaltenes [1,3,5,10]. Environmental factors, such as temperature, the presence of oxygen or other electron acceptors, nutrients, salinity, pH, water, and osmotic stress can limit the biodegradation process by inhibiting the growth of bacteria able to degrade hydrocarbons and/or reducing the bioavailability of hydrocarbons to bacterial attack [3,10,16]. Among biological factors, the bacterial diversity and their metabolic capabilities are major characteristics influencing petroleum hydrocarbons biodegradation. Many studies were carried out with pure culture or mixed bacterial consortia isolated from petroleum hydrocarbons polluted sites [2,5,6,11,17]. Mixed bacterial consortia isolated exhibited higher biodegradation rates, as compared with pure cultures, as a result of the synergistic interactions among different bacteria of the associations, which may lead to the complete degradation of toxic petroleum hydrocarbons into non-toxic end products. Some bacteria of the associations have the ability to secrete specific degradative enzymes and growth factors, while other bacteria have the ability to produce extracellular polymers, such as biosurfactants and bioemulsifiers which increase the surface area and bioavailability of hydrocarbons for biodegradation [1,2,6,11,16].
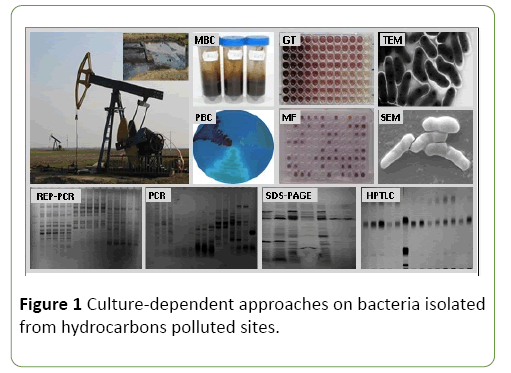
In the last decades, advances in microbiology made it possible to combine culture-dependent and culture-independent approaches in order to describe bacterial diversity and bacterial activity in petroleum hydrocarbons polluted sites [10,12,13]. The culture-dependent techniques (e.g., growth test, metabolic fingerprinting; microscopic examination by LM, TEM, SEM; biofilm formation; genetic fingerprinting by ARDRA, RAPD, REP-PCR, T-RFLP, PFGE; quantitative PCR by Q-PCR, RT-Q-PCR; protein analysis by SDSPAGE; lipid and secondary metabolites analysis by HPTLC) are useful for the isolation and characterization of novel bacterial strains which possess new degradative pathways. The culture-independent techniques (e.g., clone library method; genetic fingerprinting by ARDRA, DGGE/TGGE, LH-PCR, RAPD, RISA, SSCP, T-RFLP; DNA microarrays by 16S rRNA gene microarrays, FGA; quantitative PCR by Q-PCR, RT-Q-PCR; FISH; lipid analysis by FAME) based on direct nucleic acids (DNA, RNA) and fatty acids analysis are useful for describing community structure, and to distinguish the metabolically active members of the bacterial community [13,18]. Several culture-dependent approaches (Figure 1) were also carried out by us for the isolation and characterization of new bacterial strains of the genera Bacillus, Klebsiella, Lysinibacillus, Pseudomonas, Rhodococcus, Shewanella, Serratia, and Vibrio able to survive in the presence of different toxic petroleum hydrocarbons [19-22].
To conclude, culture-independent approaches, as well as culture-dependent approaches on pure culture or mixed bacterial consortia isolated from hydrocarbons polluted sites will contribute in the future to the development of new bioremediation strategies for environments contaminated with petroleum and petroleum products.
References
- Das N, Chandran P (2011) Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Internat 2011: 941810.
- Al-Wasify RS, Hamed SR (2014) Bacterial biodegradation of crude oil using local isolates. Inte J Bacteriol 2014: 863272.
- Leahy JG, Colwell RR (1990) Microbial degradation of hydrocarbons in the environment. Microbiol Rev 54: 305-315.
- Jain PK, Gupta VK, Gaur RK, Lowry M, Jaroli DP, et al. (2011) Bioremediation of petroleum oil contaminated soil and water. Res J Environ Toxicol 5: 1-26.
- Fathepure BZ (2014) Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front Microbiol 5:173.
- Bordenave S, Goñi-Urriza MS, Caumette P, Duran R (2007) Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat. Appl Environ Microbiol 73: 6089-6097.
- Sikkema J, de Bont JAM, Poolman B (1995) Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59: 201-222.
- Heipieper HJ, Neumann G, Cornelissen S, Meinhardt F (2007) Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl Microbiol Biotechnol 74: 961-973.
- Eberlein C, Baumgarten T, Starke S, Heipieper HJ (2018) Immediate response mechanisms of gram-negative solvent tolerant bacteria to cope with environmental stress: cis-trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl Microbiol Biotechnol 102: 2583-2593.
- Van Hamme JD, Singh A, Ward OP (2003) Recent advances in petroleum microbiology.MicrobiolMolBiol Rev 67: 503-549.
- Cerqueira VS, Hollenbach EB, Maboni F, Vainstein MH, Camargo FAO, et al. (2011) Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol 102: 11003-11010.
- Stefani FO, Bell TH, Marchand C, de la Providencia IE, El Yassimi A, et al. (2015) Culture-dependent and -independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PLoS ONE 10: e0128272.
- Widada J, Nojiri H, Omori T (2002) Recent developments in molecular techniques for identification and monitoring of xenobiotic-degrading bacteria and their catabolic genes in bioremediation. Appl Microbiol Biotechnol 60: 45-59.
- Kensa VM (2011) Bioremediation - an overview. J Ind Pollut Contr 27: 161-168.
- Koshlaf E, Ball AS (2017) Soil bioremediation approaches for petroleum hydrocarbon polluted environments. AIMS Microbiology 3: 25-49.
- Gkorezis P, Daghio M, Franzetti A,Van Hamme JD, Sillen W,et al. (2016) The interaction between plants and bacteria in the remediation of petroleum hydrocarbons: an environmental perspective. Front Microbiol 7:1836.
- Segura A, Hurtado A, Rivera B, Lazaroaie MM (2008) Isolation of new toluene-tolerant marine strains of bacteria and characterization of their solvent-tolerance properties. J Appl Microbiol 104: 1408-1416.
- Rastogi G, Sani RK (2011) Molecular techniques to assess microbial community structure, function, and dynamics in the environment. In: Ahmad I, Ahmad F, Pichtel J (eds.). Microbes and Microbial Technology: Agricultural and Environmental Applications. Springer, New York. pp: 29–57.
- Stancu MM, Grifoll M (2011) Multidrug resistance in hydrocarbon-tolerant Gram-positive and Gram-negative bacteria. J Gen Appl Microbiol 57: 1-18.
- Stancu MM (2012) Investigation of organic solvent resistance mechanisms in Vibrio alginolyticus IBBCt2. Water Air Soil Poll 223: 4495-4505.
- Stancu MM (2014) Physiological cellular responses and adaptations of Rhodococcuserythropolis IBBPo1 to toxic organic solvents. J Environ Sci 26: 2065-2075.
- Stancu MM (2016) Response mechanisms in Serratiamarcescens IBBPo15 during organic solvents exposure. Curr Microbiol 73: 755-765.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences