Research Article - (2018) Volume 8, Issue 4
Ali Mohamed Omar1, Ayman Salah Taha2* and Amal A. A. Mohamed3
1Microbiology Department, Conservation center, Grand Egyptian museum, Egypt
2Conservation Department, Faculty of Archaeology, Aswan University, Egypt
3Department of Botany, Faculty of Science, Aswan University, Aswan, Egypt
Corresponding Author:
Ayman Salah Taha
Conservation Department
Faculty of Archaeology
Aswan University, Egypt
Tel: 02 01150393937
E-mail: akinbodefoluso@gmail.com
Received Date: May 07, 2018; Accepted Date: June 18, 2018; Published Date: June 28, 2018
Citation: Omar AM, Taha AS, Mohamed AAA (2018) Microbial Deterioration of Some Archaeological Artifacts: Manipulation and Treatment. Eur Exp Biol Vol. 8 No. 3:21. doi:10.21767/2248-9215.100062
Copyright: © 2018 Omar AM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
The present study appraises an emphasis on appropriately treating of microbial deterioration of different archaeological artifacts such as papyrus, manuscripts, parchment, wood antiques and building materials.
Microbial swabs were taken from these infected artifacts and the isolated microorganisms were characterized. The following genera were identified: Aspergillus, Penicillium, Acremonium, Rhizopus, Cladosporium, Torula and Alternaria. The genus Aspergillus was the dominant genus having 49.6% of the total fungal isolates, followed by Penicillium and Acremonium.
Plant extracts were prepared from two aquatic plants, Polygonum senegalensis and Potamogeton crispus, and their antimicrobial activities against the isolated microorganisms were evaluated. Both plants showed potent antimicrobial activity.
GC-MS analysis of methanolic extracts was performed for both plants. In Polygonum senegalense, the main chemical constituent was 2-butenoic acid, 2-methyl-, dodecahydro-8- hydroxy-8a-methyl-3,5-bis(methylene)-2-oxonaphtho[2,3- b]furan-4-y (27.05%) followed by 2-cyclohexylpiperidine (10.70%), 1,1,3,3-tetramethyl-1,3-disilaphenalane (10.10%), psi,psi-carotene, 1,1',2,2'-tetrahydro-1,1'-dimethoxy- (8.50%), linoleic acid ethyl ester (6.57%) and l-(+)-ascorbic acid 2,6-dihexadecanoate (5.30%). The main chemical constituents of methanolic extract of Potamogeton crispus were 2-hydroxy-2-methyl-succinic acid, bis-(2-oxo-2-phenylethyl ester (32.70%), 2-thiazolamine, 4-(3,4- dimethoxyphenyl)-5-methyl- (15.90%), cucurbitacin B, dihydro- (8.30%) and 3-dimethylamino-2-(4-chlorphenyl)- thioacrylic acid, thiomorpholide(7.29%).
Keywords
Archiological artifacts; Microbial deterioration; Potamogeton crispus; Polygonum senegalensis
Introduction
In the last few decades, loss of cultural heritage, as a result of bio deterioration was being highly recognized. From museum objects to rock monuments, the microbial bio deterioration agents were found to be the most destructive [1]. Fungal growth on objects of cultural heritage often causes serious aesthetical spoiling due to colony formation and fungal pigments [2,3]. Fungal degradation of library materials and paintings causes different kinds of damage depending on the species of organism responsible for the attack and the characteristics of the substratum. Damage can occur because of mechanical stress, production of staining compounds or enzymatic action [4,5].
The rate of deterioration and the type of damage depend on a combination of important factors, each of which has an effect on bio deterioration, e.g., the temperature, the environmental relative humidity, moisture content of the material, light intensity and nature of the material, and microbial attack [6,7]. Museum materials such as paper, textiles and wood are also attacked by fungi causing very fast deterioration [8-11]. Most important bio deteriorative organisms of limestone are fungi because of their erosive impact and ability to penetrate inside the stone depending on the physical properties of the material [12,13]. The type of degradation caused by microorganism is usually determined by the nature of the support. Organic (wood, leather and textiles) and inorganic supports (stone, glass and metals) determines the deterioration mechanisms. For instance, fungi use the organic support itself as nutrients, while inorganic materials are transformed by several excreted metabolites which may react with the support in different ways [14,15].
Aquatic plants are a rich source for numerous bioactive compounds which exhibit antimicrobial activities [16-21].
In the present study, two dominant species of aquatic plants were used for screening their protective effect against deterioration of historical manuscripts and books caused by microbial contamination. These tested plants were Polygonum senegalensis and Potamogeton crispus belonging to families Polygonaceae and Potamogetonaceae, respectively.
Materials and Methods
Sampling
Twenty swabs of papyrus, limestone,parchment and wood antiques showing symptoms of microbial infection were collected from Grand Egyptian Museum. Illustration of these samples and their corresponding symptoms of deterioration are shown in Table 1.
| No. | Location | Sample description | No. of swabs | Symptoms |
|---|---|---|---|---|
| 1 | Grand Egyptian museum (66796) | Papyrus | 5 | white spots |
| 2 | Grand Egyptian museum (61545) | Wood | 5 | Microbial growth |
| 3 | Grand Egyptian museum (1878) | Lime stone | 5 | Black spots, |
| 4 | al-azhar library in Cairo (85265-2358) | Manuscripts | 5 | Brown spots, Black spots |
Table 1. Description of the tested archeological artifacts and their locations.
Sampling was carried out by scratching the surface of infected materials using sterilized cotton swabs and transferred right onto three prepared agar media (Dox’s agar for fungi, nutrient agar for bacteria and starch nitrate agar for actinomycetes). Plates were incubated at 28-30°C for 1-7 days depending on the microorganism (Figure 1).

Figure 1: Deterioration shape of the infected artifacts.
Identification of microbial isolates
Identification of all microbial isolates was carried out at Laboratory of Microbiology, Conservation Center, Grand Egyptian museum Egypt (Figures 2-4) [22].
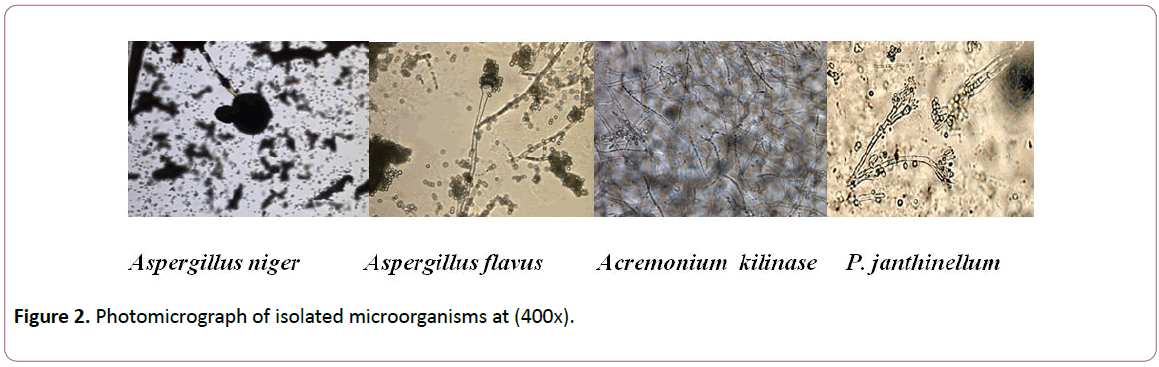
Figure 2: Photomicrograph of isolated microorganisms at (400x).
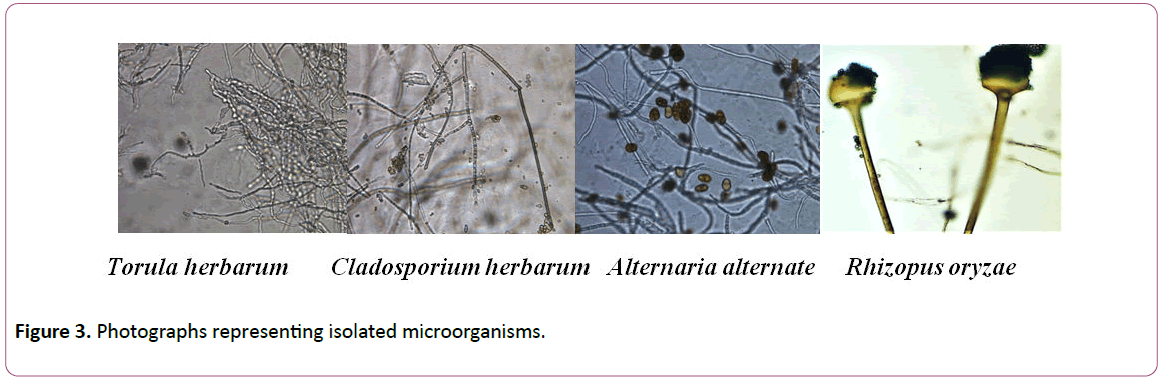
Figure 3: Photographs representing isolated microorganisms.
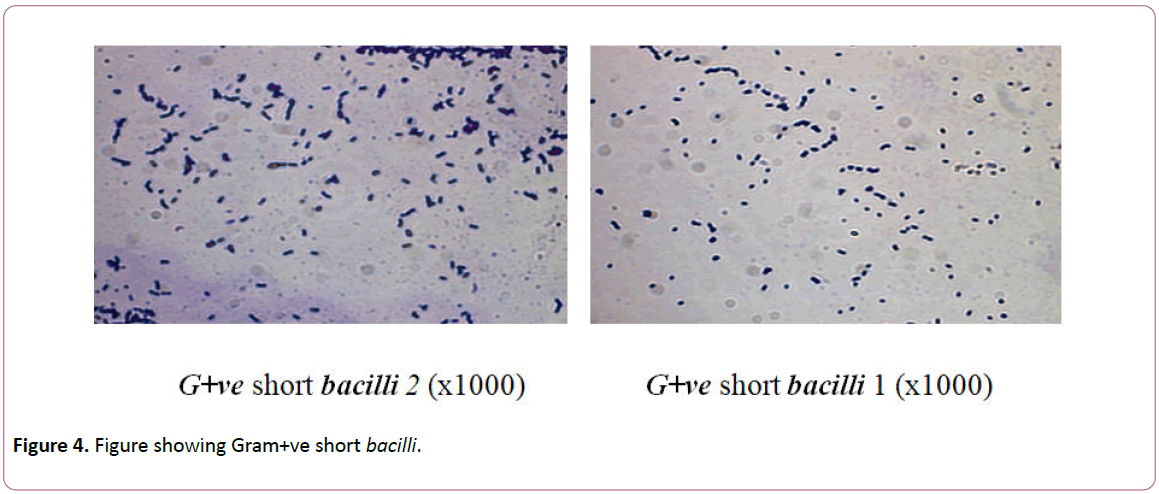
Figure 4: Figure showing Gram+ve short bacilli.
Collection and preparation of plant materials
Polygonum senegalensis (knotweed) is a widespread emergent aquatic plant, while, Potamogeton crispus (curled pondweed) are submerged aquatic perennial plants. The plants were collected from sites located (N: 24º 04.646'; E:32º 52.701').
The collected plants were authenticated and voucher specimens were sent to Herbarium. The numbers of voucher specimens are 11814 and 11815 for Potamogeton crispus and Polygonum senegalensis, respectively [23].
The plant samples were washed properly, left for air-drying and ground to a fine powder.
Extraction of plant material
For each plant, about 200 g of the ground material was extracted by methanol (100%) by soaking in the solvent, held with occasional shaking and left for overnight. The mixture was filtrated and the filtrated extract was dried under reduced pressure (~ 40ºC). Drying produced a semi-solid mass (the crude methanolic extract). The methanolic extracts of the two plants were used for antimicrobial assays.
GC/MS analysis of the extracts
The GC/MS analyses were performed at Institute of Marine Sciences, Alexandria, Egypt using Trace GC mass spectrometer IRM Calibration Status Inj Position ACQ Method. The column oven temperature was initially held at 120ºC and then increased by 5ºC/min into 200ºC with holding 2 min then increased to 280ºC (10ºC /min). The injector and detector (MS transfer line) temperatures were kept at 250ºC. Helium was used as a carrier gas at a constant flow rate of 1 ml min/1. The solvent delay was 2 min and diluted samples of 1 ml were injected automatically using Autosampler AS3000 coupled with GC in the split mode. EI mass spectra were collected at 70 eV ionization voltages over the range of m/z 40e550 in full scan mode. The ion source and transfer line temperatures were set at 200 and 250ºC, respectively. The components were identified by comparison of their retention times and mass spectra with those of WILEY 09 and NIST 11 Mass Spectral (2011) databases.
Determination of minimal inhibitory concentration (MIC) of plant extracts against the isolated microorganisms
Gradient concentrations of each plant extract ranged from 400 to 3000 ppm were prepared by diluting from their respective stock solutions.
One ml of bacterial isolate suspension was spread onto nutrient agar plate and for fungi, one ml of spore suspension was spread on Dox’s agar plate. Plates were allowed to dry then a cork pourer was used to make three pores in each plate. In each pore, 100 μl of each concentration (from 400 to 3000 ppm) of the tested extracts. Plates were incubated at 30 oC for 1-3 days comparing with Control plates (ethyl alcohol instead of extracts). The minimal inhibitory concentration (MIC) was determined by measuring the inhibition zone [14].
Minimal inhibitory concentration (MIC) test is considered the “gold standard’’ for determining the susceptibility of organisms to particular antimicrobial substance and are therefore used to judge the performance of all other methods of susceptibility testing.
The MIC can be defined as “the lowest concentration of the antimicrobial agent that completely inhibits visible growth of a tested organism on a solid medium around a disk containing that particular concentration as judged by the naked eye” (after overnight incubation for bacteria and after 1-3 days for fungi).
Result and Discussion
Microbial isolates
Twenty swabs were taken from four sites and their preliminary classification by plate count resulted in 40 colonies of fungi isolated on Dox’s agar and 13 colonies of bacteria isolated on nutrient agar. No colonies of actinomycetes were detected on the starch nitrate agar.
Identification of the microbial isolates
The resulted microbial colonies were subjected to preliminary characterization depending on the type of organism as mentioned previously. The following genera were identified: Aspergillus, Penicillium, Acremonium, Rhizopus, Cladosporium, Torula and Alternaria (Table 2). Fro m the results, it can be seen that the genus Aspergillus was the dominant genus in four objects having 49.6% of the total fungal isolates, followed by Penicillium and Acremonium.
| Sample | Total fungal colonies | A. niger | A.flavus | P. janthinellu | Acremonium Kiliasee | Torula herbarum | Rhizopus oryzae | Claod. Herbarum | Alternaria alternata | G+ve short Bacillus 1 | G+ve short bacillus 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Papyrus | 9 | 9 | ND | ND | ND | ND | ND | ND | ND | ND | 4 |
| Wood | 10 | 2 | ND | 3 | ND | 4 | 1 | ND | ND | 2 | ND |
| Lime stone | 16 | 3 | 2 | ND | 3 | ND | 2 | 3 | 3 | 2 | 2 |
| Manuscripts | 5 | 3 | 2 | ND | ND | ND | ND | ND | ND | 2 | 1 |
| 40 | 17 | 4 | 3 | 3 | 4 | 3 | 3 | 3 | 6 | 7 |
Table 2. Identification of the fungal and bacterial species isolated from archaeological artifacts.
Isolates of each genus were subjected to identification based on morphological characteristics. Results showed that isolates of Aspergillus were A. niger and A. flavus,isolates of Penicillium were identified as P. janthinellum, isolates of Acremonim were identified to be A. kilinese and Isolates of other genera were identified as Rhizopus oryzae, Cladosporium herbarum, Torula herbarum and Alternaria alternate (Table 2).
The highest percentage of the fungal strains was recorded for genus Aspergillus niger (32%) followed by A. flavus and Torula herbarum (7.5%) and other genera Rhizopus, Penicillium, Cladosporium, Acremonium and Alternaria (5.6%).
The isolated fungal strains are considered asthe most biodeteriorates for cultural and historical heritage [1]. The results are in accordance with old findings, who isolated Aspergillus flavus, Alternaria alternata,Pencillium chrysogenum, Rhizopus and other fungal strains from archeological artifacts in the museum environment [5,6,8].
In the present study, the most prevalent fungus detected in all infected artifacts was Aspergillus niger. In the same context, Arroyo reported Aspergillus, Penicillium and Cladosporium as the most molds growing in indoor environments(Table 2).
Isolates of bacteria genera were identified as G+ve short bacilli1 and G+ve Short bacilli2. For bacterial isolates, members of genus G+ve short bacilli1 represented 11% followed by G+ve Short bacilli2 of 13.2% (Table 2). These isolated spore-forming bacteria are prevailing and frequently been detected on historical articles [15].
In the present study, it was apparent that limestone is the more susceptible artifact for microbial damage. This result is in line with that the limestone environments are influenced by decay of soft-rot fungi due to alkaline conditions [24].
Determination of minimal inhibitory concentration (MIC) of plant extracts against the isolated microorganisms
Table 3 shows the antimicrobial assay results of Polygonum Senegalense; 3000 ppm was the MIC that inhibited all the isolates where the diameter of inhibition zone ranged between (20-40 mm).
| Microbial isolate | Mean diameter of inhibition zone (mm) at different concentrations (ppm) of Polygonum senegalense | |||||
|---|---|---|---|---|---|---|
| 400 | 600 | 800 | 1000 | 2000 | 3000 | |
| Aspergillus niger | 0 | 0 | 20 | 25 | 35 | 40 |
| Aspergillus flavus | 0 | 0 | 0 | 19 | 21 | 26 |
| Acremoniumkilinase | 0 | 0 | 0 | 0 | 19 | 23 |
| Torula herbarum | 0 | 0 | 0 | 0 | 20 | 24 |
| Cladosporium herbarum | 0 | 0 | 0 | 0 | 20 | 25 |
| Alternaria alternate | 0 | 0 | 0 | 0 | 22 | 27 |
| P. Janthinellum | 0 | 0 | 0 | 19 | 23 | 27 |
| Rhizopus oryzae | 0 | 0 | 0 | 0 | 19 | 22 |
| G+ve short bacilli 1 | 0 | 0 | 0 | 0 | 0 | 20 |
| G+ve short bacilli 2 | 0 | 0 | 0 | 0 | 0 | 23 |
Table 3. Determination of inhibition zone (mm) of fungal isolates treated by different concentrations of Polygonum senegalense.
Table 4 shows the antimicrobial assay results of Potamogeton crispus. The MIC that inhibits all the tested fungal isolates was 2000 ppm and the mean diameter of inhibition zone ranged between 18-41 mm.
| Microbial isolate | Mean diameter of inhibition zone (mm) of different concentrations (ppm) of Potamogetoncrispus | |||||
|---|---|---|---|---|---|---|
| 400 | 600 | 800 | 1000 | 2000 | 3000 | |
| Aspergillus niger | 0 | 20 | 25 | 30 | 35 | 40 |
| Aspergillus flavus | 0 | 19 | 26 | 30 | 36 | 41 |
| Acremoniumkilinase | 0 | 0 | 0 | 0 | 19 | 24 |
| Torula herbarum | 0 | 0 | 0 | 0 | 20 | 25 |
| Cladosporium herbarum | 0 | 0 | 0 | 0 | 18 | 23 |
| Alternaria alternate | 0 | 0 | 0 | 20 | 27 | 35 |
| P. Janthinellum | 0 | 0 | 0 | 19 | 25 | 30 |
| Rhizopus oryzae | 0 | 0 | 0 | 20 | 28 | 33 |
| G+ve short bacilli 1 | 0 | 0 | 23 | 29 | 32 | 35 |
| G+ve short bacilli 2 | 0 | 0 | 0 | 19 | 25 | 33 |
Table 4. Determination of inhibition zone (mm) of fungal isolates treated by different concentrations of Potamogeton crispus.
Chemical composition of the extracts
GC-MS analysis of methanolic extract of Polygonum senegalense shown in Table 5 revealedthat the main chemical constituent was the organic compound 2-Butenoic acid, 2- methyl-,dodecahydro-8-hydroxy-8a-methyl-3,5- bis(methylene)-2-oxonaphtho[2,3-b]furan-4-y (RT=18.73 min, 27.0 %) followed by 2-Cyclohexylpiperidine (RT=19.31 min, 10.70%), 1,1,3,3-Tetramethyl-1,3-disilaphenalane (RT=20.85 min, 10.10%), psi.,.psi.-Carotene, 1,1',2,2'-tetrahydro-1,1'- dimethoxy- (RT=14.02 min, 8.50%), Linoleic acid ethyl ester (RT=31.71 min, 6.57%) and l-(+)-Ascorbic acid 2,6- dihexadecanoate (RT=31.44 min, 5.30%). These compounds were involved in antimicrobial activities [18,20, 21].Polygonum senegalensehas been reported to show potent antimicrobial activity [25-28].
| Compound name | Retention time | Percentage (%) |
|---|---|---|
| α-D-Glucopyranoside, methyl 2-(acetylamino)-2-deoxy-3-O-(trimethylsilyl)-, cyclic butylboronate | 12.05 | 3.28 |
| 8,14-Seco -3,19-epoxyandrostane-8,14-dione,17-acetoxy-3ß-methoxy | 12.28 | 2.05 |
| 17-acetoxy-3β-methoxy-4 | 12.58 | 2.47 |
| psi.,.psi.-Carotene, 1,1',2,2'-tetrahydro-1,1'-dimethoxy- | 14.02 | 8.5 |
| 2-Butenoic acid, 2-methyl-, dodecahydro-8-hydroxy-8a-methyl-3,5-bis(methylene)-2-oxonaphtho[2,3-b]furan-4-y | 18.73 | 27.05 |
| 2-Cyclohexylpiperidine | 19.31 | 10.7 |
| 2(5H)-Furanone | 20.08 | 3.23 |
| 1,1,3,3-Tetramethyl-1,3-disilaphenalane | 20.85 | 10.1 |
| 2,5-Bis[(trimethylsilyl)oxy]pyrazin | 21.65 | 2.56 |
| Phenol, 2-(1-methylpropyl) thio- | 24 | 5.4 |
| 4-Decenoic acid, methyl ester | 24.15 | 2.93 |
| (+-)-5,5-Dimethyl-4-(3-oxobutyl)dihydro-2(3H)-furanone 4-(2,4-dinitrophenylhydrazone) | 24.38 | 3.26 |
| Fenretinide | 25.28 | 3.02 |
| 17-Octadecynoic acid | 29.82 | 1.38 |
| l-(+)-Ascorbic acid 2,6-dihexadecanoate | 31.44 | 5.3 |
| Linoleic acid ethyl ester | 31.71 | 6.57 |
| 1,2,4-Trioxolane-2-octanoic acid, 5-octyl-, methyl ester | 33.39 | 0.76 |
Table 5: Chemical constituents identified in the methanolic extract of Polygonum Senegalense.
Table 6 shows GC-MS analysis of methanolic extract of Potamogetoncrispus. The main chemical constituents were 2- Hydroxy-2-methyl-succinic acid, bis-(2-oxo-2-phenyl-ethyl ester (RT=5.69 min, 32.70%), 2-Thiazolamine, 4-(3,4-dimethoxyphenyl)-5-methyl (RT=11.68 min, 15.90%), Cucurbitacin B, dihydro- (RT=14.18 min, 8.30%) and 3- Dimethylamino-2-(4-chlorphenyl)-thioacrylic acid, thiomorpholide (RT=18.94 min, 7.29%). It is likely that the bioactive compound Cucurbitacin may be involved in biological activity of other compounds [29-33]. As indicated from previous studies, extracts of Potamogetoncrispus showed forceful antibacterial and antifungal activities [34].
| Compound name | Retention time | Percentage (%) |
|---|---|---|
| 2-Hydroxy-2-methyl-succinic acid, bis-(2-oxo-2-phenyl-ethyl ester | 5.69 | 32.7 |
| 2-Thiazolamine, 4-(3,4-dimethoxyphenyl)-5-methyl- | 11.68 | 15.9 |
| 2-(1-Methyl-1-silacyclobutyl)benzoic acid trimethyl-silyl ester | 11.82 | 4.13 |
| 3,5,6-Trimethyl-p-quinone, 2-(2,5-dioxotetrahydrofuran-3-yl)thio- | 12.01 | 1.5 |
| β-N-Acetylneuraminic acid, | 13.39 | 2.8 |
| Cucurbitacin B, dihydro- | 14.18 | 8.3 |
| Carbamic acid, N-methyl-N-[6-iodo-9-oxabicyclo[3.3.1]nonan-2-yl]-, ethyl ester | 15.06 | 5.44 |
| α-D-Glucofuranose, 6-O-(trimethylsilyl)-, cyclic 1,2:3,5-bis(butylboronate) | 17.15 | 4 |
| 3-Dimethylamino-2-(4-chlorphenyl)-thioacrylic acid, thiomorpholide | 18.94 | 7.29 |
| 2-Bromotetradecanoic acid | 19.29 | 4.23 |
| Digitoxin | 20.08 | 3.05 |
| Disilane, 1,1,1,2,2-pentamethyl-2-[(cyclopropyl)(phenylthio)methyl]- | 20.85 | 3.56 |
| 4-Piperidineacetic acid, 1-acetyl-5-ethyl-2-[3-(2-hydroxyethyl)-1H-indol-2-yl]-α-methyl-, methyl ester | 21.61 | 3.21 |
| Erucic acid | 24 | 3.06 |
Table 6: Chemical constituents identified in the methanolic extract of Potamogetoncrispus.
Conclusion
It can be concluded that Polygonum senegalensis and Potamogeton crispus are rich sources of biologically active compounds. The present study provided adequate records on antimicrobial activity and biochemical composition of the two plants.
The present study suggests that the microbial deterioration of archaeological artifacts could be avoided by using extracts of these aquatic plants at their lowest concentration, i.e. MIC, promising a future scope to protect cultural heritage [35-37].
Acknowledgment
The authors of this paper are thankful to the Microbiology laboratory in conservation center in the Grand Egyptian Museum for helping and providing necessary facilities for this research work, Grateful thanks to the staff members of Al-Azhar library in Cairo for their kind assistance and cooperation for samples collection.