Research Article - (2020) Volume 10, Issue 3
Juby Elsa Joseph and Veena Gayathri Krishnaswamy*
Department of Biotechnology, Stella Maris College, Chennai, Tamil Nadu, India
Received date: June 2, 2020; Accepted date: June 13, 2020; Published date: June 22, 2020
Citation: Joseph JE, Krishnaswamy VG (2020) Isolation of Natural Rubber Latex Degrading Bacterial Consortium from Rubber Plantation Area. Eur Exp Biol Vol.10 No.3:9. doi:10.36648/2248-9215.10.3.106
The huge quantity of waste rubber materials can cause environmental problems that are the accumulation of recalcitrant organic compounds in the environment which need great concern, sufficiently long (persistence) that they have undesirable effects. Most commonly-used fibers and natural polymers are natural rubber, especially starches, gelatin, wood and cotton are inherently biodegradable under favourable conditions although the intensity of degradation decreases with the increasing molecular weights of the polymers. Natural rubber (NR) is both a photo synthetically-renewable resource and an environmentally degradable material. It is an unsaturated high molecular weight polymer, and in nature, is expected to degrade very slowly in comparison with other natural polymers. Various physical, chemical and biological methods have been proposed to solve this problem. NR-degrading bacteria are widely distributed in soil, water and sewage and interestingly, with some exceptions, belong to the actinomycetes. As a group, the bacteria and actinomycetes are characterized by an ability to digest complex materials and to use unusual molecules and are thus significant decomposers in soils. Present review focuses on the degradation of natural rubber by different microbes involved in degradation, analysis of degradation products and biodegradation pathway for technically important polymers. This review may provide an opportunity for further studies about the applications of biotechnological processes used as a tool for rubber waste management.
Actinomycetes; Bacteria; Biodegradation; Natural rubber
Latex is a naturally occurring polymer of isoprene often known as cis-1,4-polyisoprene with a molecular weight of 100,000 to 1,000,000. Some natural rubber sources like guapercha are composed of trans-1,4-polyisoprene, a structural isomer that has similar, but not identical, properties. Polyisoprene can also be created synthetically, producing what is called synthetic natural rubber, while the natural and synthetic routes are completely different. Natural Rubber Latex (NRL) is a specific type of rubber derived from milky sap of the Hevea brasiliensis tree that is refined and treated to form the raw liquid latex used to make gloves. It is also used to manufacture numerous diverse products namely tyres, clothing, medical products and toys. Currently the rubber is harvested mainly in the form of the latex from some trees. This natural rubber latex is a sticky, milky colloid drawn off by making incisions into the bark and collecting the fluid in specific vessels. This process is known as "tapping". The latex is then refined to rubber ready for commercial processing and a solution of ammonia can be used to prevent the coagulaon of raw latex while it is being transported from its collection site [1].
Natural Rubber (NR) contains a minimum of 90% rubber hydrocarbon together with small amounts of proteins, resins, fatty acids, sugars, and minerals [2]. The main constituent of natural rubber is cis-polyisoprene. Rubber particles are found to be surrounded by a single membrane of phospholipid with hydrophobic tails pointed inward.
Once the rubber is vulcanized, it will be turned into a thermoset and most of the rubber in everyday used to be vulcanized to a point where it shares properties of both; i.e., if it is heated and cooled, it is degraded but not destroyed. This natural rubber latex is also a natural, biodegradable product, containing no petroleum by-products or dioxins that enables disposal by either land fill or incineration without environmental damage. Organic impurities present in the rubber can support microbial growth as well [3,4]. There are several evidences that these natural rubbers that can be degraded by the number of fungi and bacteria present in the soil [5-11].
Since accumulation of rubber waste in the environment is becoming a worldwide problem [12], it is worth trying to develop a microbial remedy for the disposal of natural rubber waste [13]. The main constituent of natural rubber is a compound called cis, 1-4 polyisopropene which is relatively resistant to microbial decomposition when compared with many other natural polymers. At the same time, there were many reports published before implying the application of microorganisms in the treatment of the effluents of natural rubber latex [14]. The extent, vigor and rate of microbial degradation are influenced by the factors like the rubber formulation, the microorganism present and their interaction with its surrounding environment. Rubber wastage and the disposal of waste tires have become two major challenges of rubber industry that leads to much environmental pollution. Thus, numerous remedies of physical, chemical and biological methods have been proposed to solve this problem.
Bacteria can use and degrade natural rubber latex as the sole source of carbon and energy were isolated from different ecosystems. About 42 rubber-degrading bacteria were isolated and all rubber-degrading isolates were identified as a member of Actinobacteria which is a Gram-positive bacteria characteristic of forming large group of mycelia. Interestingly no Gram-negative bacteria could be isolated. The growing problem of environmental pollution caused by synthetic plastics has led to the search for alternative materials like biodegradable and renewable plastics. Of the biopolymers presently under development, starch or natural rubber is one of the promising alternatives. This problem has extensively triggered research into finding and producing alternative synthetic materials and recent researches widely search for multipurpose, versatile, biodegradable plastics [15,16].
Collection of latex material
The liquid latex sample was collected from Kerala rubber plantation area (Kottayam). Ammonia solution was used to prevent the coagulation of raw latex while it is being transported from its collection site.
Sample collection
The soil sample was collected from Kerala rubber plantation area. They were collected from the rooting part of the rubber trees.
Media used
A mineral salts medium was used for the acclimatization of the bacterial consortium (Table 1) [17].
| Media composition | g/L |
|---|---|
| di-Potassium hydrogen phosphate (K2HPO4) | 1 |
| Magnesium sulphate (MgSO4.7H20) | 0.5 |
| Potassium Nitrate (KNO3) | 1 |
| Distilled water | 1 L |
| pH | 7 |
Table 1: Media composition.
Acclimatization
Prepared 100 ml mineral salts medium each in two different conical flasks. Then to one of the conical flasks 1 ml of preserved latex solution was added and to the other conical flask 0.5 ml of the same latex solution with 0.10% of yeast extract as nitrogen source. Then they were sterilized in the autoclave. After sterilization once the media was cooled to each of them 5 grams of the collected soil sample was inoculated. Then it was allowed to grow or kept for incubation for 2 days.
After the incubation the microbial growth was observed under microscope. So, it was found that the growth of microbes in the medium that was inoculated with 1 ml of latex solution is higher. Then both the cultures containing 0.5 ml latex as well as 1 ml latex were pelletized at 10,000 rpm for 10 minutes. The supernatant was discarded, and the pellets were dissolved in a sterilized freshly prepared 100 ml of mineral salts medium with 0.10% yeast extract as a nitrogen source to enhance bacterial growth. Figure 1 shows the pelletized bacterial culture solution.
Isolation of different types of bacterial isolates from the mixed consortium
Using the pelletized culture solution serial dilution was done was spread plated on sterile nutrient agar plates from the dilutions 10-5, 10-6 and 10-7. Then the different types of individual isolated bacterial colonies were taken based on their morphological characters and streaked on other freshly prepared nutrient agar slants in sterile screw cap tubes. These bacterial slants were preserved for further experiments.
Determination of the molecular weight of the NR-latex solution
For more accurate evaluation of the average Molecular Weight (MW) of the collected NR-latex solution a capillary Ubbelohde viscometer was used. The collected NR-latex solution was diluted with distilled water at different concentrations like 0.01, 0.02, 0.03, 0.04, 0.05, 006, 0.07, 0.08, 0.09 and 0.10. Then according to the time taken by each of those samples to reach their specific end point in the viscometer (Table 2), the specific viscosity was calculated for each sample (Table 3). To find out specific viscosity the given equation was used, where t0- the time taken by water and ts-time taken by the different diluted latex sample.
| Sample conc. | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.07 | 0.08 | 0.09 | 0.1 | Dist. water |
| Time taken (sec.) | 110 | 100 | 90 | 80 | 78 | 75 | 83 | 87 | 90 | 92 | 72 |
Table 2: Time taken by each sample.
| Sample | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.07 | 0.08 | 0.09 | 0.1 |
| Specific viscosity | 0.527 | 0.388 | 0.25 | 0.11 | 0.083 | 0.042 | 0.152 | 0.21 | 0.25 | 0.28 |
Table 3: Specific viscosity of each sample.
Specific viscosity, 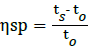
From these specific viscosities of the individual samples a graph was plotted, and the intrinsic viscosity was calculated (Figure 2).
Using this intrinsic viscosity value, the molecular weight of the latex was calculated with help of an equation given below that includes Mark-Houwink constants K and α. According to Robert O Ebewele [18],
Intrinsic viscosity, η = KMα
Determining the mineralization rate of NRlatex by the mixed microbial consortium
The percentage mineralization rate taken place at each concentration on each day was determined with the help of titration method and growth OD. Biodegradation of the poly (cis-1,4-isoprene) and poly (trans-1,4-isoprene) hydrocarbon chains to carbon dioxide can be obtained by determining the respiratory CO2 released during cultivation of cells in presence of polyisoprene as sole carbon source [19]. Determination was carried out in tightly closed glass bottles and boiling tubes by using the property of Ba(OH)2 to precipitate CO2 as BaCO3. The bottles, containing 50 ml of mineral salts medium, 0.10%-0.50% of milled poly (cis- 1,4-isoprene) or poly (trans- 1,4-isoprene), and the boiling tubes containing 15 ml of 0.2 M Ba(OH)2 solution inoculated with 1 ml of the pelletized culture grown for 48 h. Consumption of HCl for precipitation of CO2-3 as BaCO3 was determined for each period by titrating with 0.1 M HCl according to the following equation, yielding the percentage mineralization.
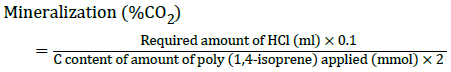
As indicator, phenolphthalein was added, and the end point of titration was determined by alteration of the colour from magenta to uncoloured. The growth OD value for each of the concentrations on each day was also taken. This growth OD value was measured with the help of UV-spectroscopy. It was done by diluting each sample with distilled water at 1:10 ratio.
Determining the % mineralization rate at different concentration
In sterilized boiling tubes 15 ml of 0.2 M Ba(OH)2 was poured. Then sterilized bottles were taken and freshly prepared mineral salts medium of 50 ml was added to each of them. To each of the 5 set of bottles containing mineral salts medium add 0.05 μl, 0.1 μl, 0.15 μl, 0.2 μl and 0.25 μl of NRsolution latex solution indicating 0.10%, 0.20%, 0.30%, 0.40% and 0.50% respectively. These bottles containing medium along with latex was sterilized. Followed by which 1 ml of the pelletized culture solution was added to each of those bottles. These bottles were connected to each boiling tube containing the Ba(OH)2 solution with sterilized plastic tubes and they were properly sealed. Then these boiling tubes containing Ba(OH)2 solution was also properly sealed. Then they were kept for overnight incubation. Since for each concentration there were 5 culture bottles, each culture was assorted for determining each day’s mineralization rate from day 1 to day 5 continuously. After incubation their mineralization rate was determined with the help of titration method. Thus, it was possible to find out at which concentration maximum mineralization takes place (Figure 3).
Determining the best carbon and nitrogen (C and N) source for the bacterial consortium
To the sterilized glass bottles, 0.10% of latex was added along with 50 ml mineral salts medium in each bottle. To each of those bottles 500 mg of C and N sources like glucose, lactose, sucrose, (C-sources) beef extract, yeast extract, ammonium sulphate and ammonium nitrate (N-source) were added respectively. These bottles were sterilized and 1 ml of pelletized bacterial culture was added to each of the bottles. They were connected to the sterilized boiling tubes containing 15 ml of 0.2 M Ba(OH)2 solution with sterile plastic tubes and were properly sealed. Then it was kept for overnight incubation. After the 24 h of incubation the % mineralization rate for each of the C and N source were calculated by the same titration method. Then the growth OD of each of those cultures was also measured with the help of a UVspectroscopy. Thus, it was possible to determine which carbon and nitrogen source helps in the maximum mineralization.
Determining the % of mineralization rate of NR by individual bacterial isolates
About 50 ml of Mineral Salts Medium (MSM) was poured to sterilized bottles. The 0.10% of the NR-latex solution was added to each those bottles and autoclaved. Then 1ml of the individual bacterial isolates was inoculated to each of the 5 set of bottles. Each bottle was connected to boiling tubes containing 15 ml Ba(OH)2 solution and kept for overnight incubation. The mineralization rate by each of those bacteria was determined by the titration method. Growth OD value was also measured on each day with the help of the UV spectroscopy. Thus, by titration method it was possible to determine which bacterial culture gives the maximum mineralization or degradation of the latex compound.
Biochemical characterization for morphological identification of bacterial isolates
All the isolated bacterial strains were studied through initial tests like motility test and gram staining. The morphological and biochemical characterization of the bacterial isolates were done with the help of tests like IMViC test, oxidase test, carbohydrate fermentation test (glucose, sucrose, raffinose), triple sugar iron agar test, catalase test, lipid hydrolysis, phenylalanine deaminase test and urease test. Antibiotic sensitivity of bacterial species was tested by Kirby Bauer’s procedure to find out the resistant pattern to various antibiotics like ampicillin, ciprofloxacin, tetracycline, erythromycin, nalidixic acid and norfloxacin. The sensitivity and resistance of each of the bacterial strains were determined.
Screening and isolation of bacterial consortium from soil sample
In the mineral salts medium, which was inoculated with 5 g of the collected soil sample and the liquid NR-latex, microbial growth was observed when visualized under light microscope. This bacterial consortium was pelletized and after platting it on nutrient agar individual bacterial isolates were obtained as Figure 4 shows and they were preserved on nutrient agar slants.
Average molecular weight of latex solution
The latex solution was diluted with distilled water at different concentrations and their specific viscosity was calculated. With the calculated specific viscosity of each sample a graph was plotted. From that graph, intrinsic viscosity was found out by extending the straight line to the y-axis in the graph. Thus, the intrinsic viscosity (η) was equal to 0.66.
By substituting the value of intrinsic viscosity in the equation given below, the average molecular weight of the liquid latex was found.
Intrinsic viscosity, η = KMα
Thus, the average molecular weight of the natural rubber latex was 45,184.08.
Mineralization of different concentration of latex by the bacterial consortium
Based on the mineralization percentage and growth of bacterial consortium a graph was plotted (Figure 5). In the present study, effect on latex solution at different concentrations (0.10%, 0.20%, 0.30%, 0.40%, and 0.50%) by the bacterial consortium was examined. From the graph, it was found that maximum mineralization of latex happens at 0.10% concentration. The maximum mineralization took place on day 5 in the culture that contained 0.10% concentration of latex giving maximum value of 5.98 × 10-8. The maximum growth of the bacterial consortium was seen on the third day in 0.10% concentration of latex culture solution and the lowest growth was seen in the 0.40% concentration on fourth day giving values of about 0.997 and 0.050 respectively. While the growths of bacterial culture at other latex concentrations like 0.20%, 0.30%, 0.50% was giving different growth values. From the graph, on day 3 the growth of bacterial consortium was maximum. When the growth OD graph was compared with the mineralization graph it was found that the mineralization rate has a sudden increase in all the culture solution having different concentration of latex solution.
Likewise, the carbon and nitrogen source which can enhance the mineralization rate was thus determined by the same method.
Mineralization of 0.10% latex with different nitrogen sources
Different nitrogen sources were added to the mineral salts medium for the enhancement of bacterial growth. From the experiment it was found that the maximum mineralization was taken place on day 5 in the culture solution that was treated with yeast extract giving a value 6.14 × 10-8. While the growth OD value was giving maximum value in the culture solution treated with beef extract on day 3 giving a value about 0.297. These growth OD values were observed with the help of UVspectroscopy at 660 nm. Figure 6 shows that each of the cultures having different nitrogen sources like beef extract, yeast extract, ammonium sulphate and ammonium nitrate as their nitrogen source have an initial bacterial growth. Their growth later decreased but gradually starts increasing again. The culture having beef extract showed its maximum growth on day 3. Yeast extract also showed increased growth of the bacterial consortium of about 0.288 on fifth day. Ammonium nitrate showed the lowest growth of bacterial consortium on day 5 giving a value about 0.092. In the same way the carbon source which can enhance the mineralization was also determined.
Mineralization of 0.10% latex with different carbon sources
The mineralization of latex and the growth of the bacterial consortium by the different carbon sources were determined with the help of a graph. Based on the Figure 7, it was found that maximum mineralization was taken place on day 4 that was treated with sucrose giving a value 5.89 × 10-8. The maximum growth of bacterial consortium was seen on day 1 giving a value 0.315 that was treated with glucose as the carbon source. In case of the growth of bacterial consortium by the different carbon sources it was found that sucrose had the maximum growth followed by lactose and then glucose. Their growth had a sudden increase initially but gradually it was decreasing.
Mineralization of natural rubber by individual bacterial strains
Based on the Figure 8, it was observed that the bacterial isolate that was named J-3 was able to do maximum mineralization. J-3 has done its maximum mineralization on day 5 giving a value of about 6.31 × 10-8. In the initial stage all the bacterial isolates showed a sudden increase in mineralization of latex. But there was a gradual decrease followed by an increase again on day 5. Likewise, the growth OD value of each individual isolates on each day was also measured. The growth was highest in the bacterial isolate that was named J-4 on day 5. In case of J-1, J-3 and J-5, the growth was increasing day by day in a slow rate. But in case of J-2, J-4 and J-6 there was an increase in their growth initially followed by a decrease and a slight increase later.
Characterization of the bacterial strains present in the consortium
The bacterial isolates were characterized according to their morphological characteristics. Further characterization of these individual bacterial isolates was done by carrying out gram staining, motility test and different biochemical tests like IMViC test, oxidase test, carbohydrate fermentation test (glucose, sucrose, raffinose), Triple Sugar Iron agar test (TSI), catalase test, phenylalanine deaminase test and urease test. The antibiotic sensitivity test was performed as well for different antibiotics like ampicillin, ciprofloxacin, tetracycline, erythromycin, nalidixic acid and norfloxacin.
In the Table 4 it is given that by gram staining it was found that only J-6 was gram positive rods and all other isolates were gram negative rods. By hanging drop method, it was found that all the six isolated bacteria were motile. In case of biochemical test, it was found that in indole test except J-5 all other bacterial isolates were positive. In methyl-red test except J-2 and J-5 all others were positive while in Voges- Proskauer test all the isolates gave negative results. In citrate utilization test it was found that only J-6 can utilize citrate giving a positive result. In oxidase test all the bacterial isolates gave positive results. In case of carbohydrate fermentation test, only J-6 gave negative results for sucrose and raffinose and J-4 gave negative results for glucose. For catalase test all the isolates responded positively. In phenylalanine deaminase test every isolate gave negative results while in urease test all the isolates responded positively. In case of TSI agar test, every isolate gave acid butt and alkaline slant but only J-6 had gas production as well.
| Test | J-1 | J-2 | J-3 | J-4 | J-5 | J-6 |
|---|---|---|---|---|---|---|
| Gram staining | Gram negative rods | Gram negative rods | Gram negative rods | Gram negative rods | Gram negative rods | Gram positive rods |
| Motility test | + | + | + | + | + | + |
| Indole test | + | + | + | + | - | + |
| Methyl-red test | + | - | - | + | - | + |
| Voges-proskauer test | - | - | - | - | - | - |
| Citrate Utilisation test | - | - | - | - | - | + |
| Oxidase test | + | + | + | + | + | + |
| Carbohydrate fermentation test | G* (+) | G (+) | G (+) | G (-) | G (+) | G (+) |
| S* (+) | S (+) | S (+) | S (+) | S (+) | S (-) | |
| R* (+) | R (+) | R (+) | R (+) | R (+) | R (-) | |
| Triple Sugar Iron agar test | acid butt and alkaline slant | acid butt and alkaline slant | acid butt and alkaline slant | acid butt and acid slant | acid butt and alkaline slant | acid butt and alkaline slant with gas production |
| Catalase test | + | + | + | + | + | + |
| Phenylalanine deaminase test | - | - | - | - | - | - |
| Urease test | + | + | + | + | + | + |
Table 4: Biochemical Tests for the six bacterial strains.
With the help of Bergey ’ s Manual of Systematic Bacteriology, genus level identification of the six bacterial isolates was accomplished. Thus, it was found that J-1, J-2, J-3, J-5 can be assumed to be an Aeromonas sp., and J-4 found to be Pseudomonas sp. while J-6 can be assumed as Bacillus sp. Among the six different isolates, four of them (J-1, J-2, J-3, J-5) were assumed to be Aeromonas sp., (J-4) was assumed to be Pseudomonas sp. and (J-6) was assumed to be Bacillus sp.
Antibiotic sensitivity test
In the Table 5 it was given that in antibiotic sensitivity test, ciprofloxacin, tetracycline, nalidixic acid and norfloxacin are sensitive to all the individual bacterial strains.
| Antibiotic disc used | J-1 | J-2 | J-3 | J-4 | J-5 | J-6 |
|---|---|---|---|---|---|---|
| Ampicillin | Sensitive | Sensitive | Sensitive | Resistant | Sensitive | Resistant |
| Ciprofloxacin | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive |
| Tetracycline | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive |
| Erythromycin | Sensitive | Sensitive | Resistant | Resistant | Sensitive | Resistant |
| Nalidixic acid | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive |
| Norfloxacin | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive |
Table 5: Antibiotic sensitivity test.
Ampicillin was resistant to the bacterial strain named J-4 while sensitive to all other bacterial strains. Erythromycin was resistant to J-3 and J-4 but sensitive to the other bacterial strains.
Rubber compounds are important group of contaminants, for which efficient treatment methods are required. Biodegradation constitutes the primary mechanism for contaminant removal. However, researches were conducted to isolate polyisoprene rubber (latex gloves) degrading microorganisms from soil. Twelve bacterial strains were isolated through enrichment technique and were screened for their capability to degrade rubber. Best growth was observed with Bacillus sp. S10 [20]. Likewise, in the present study, the natural rubber (latex solution) degrading microbes were isolated from soil. Six bacterial strains were isolated through enrichment technique and were screened for their capacity to degrade rubber in the liquid state. The culture assumed as Aeromonas sp. i.e. J-3 gave the highest mineralization % of natural rubber latex giving value about 6.31 × 10-8 within 5 days.
Warneke et al. [19] isolated axenic cultures capable of degrading poly (trans-1,4-isoprene) or Gutta Percha (GP) of which six bacterial strains were capable of utilizing synthetic poly (trans-1,4-isoprene) as sole carbon and energy source for their growth. In cultures with GP grains of 120 μm-250 μm, 40% of the substrate was metabolized to CO2 within 78 days, while only 25% of the poly (trans-1,4-isoprene) granules of 1250 μm-1400 μm was mineralized in the same period by N. takedensis WE30. But in the present study as a carbon source natural rubber latex solution was used instead of solid latex material for the easy and faster mineralization by the bacterial strains. The bacterial consortium isolated in the present study was able to mineralize the latex up to 0.50%, in which optimum degradation was obtained in 0.10% within 5 days of incubation.
The microbial susceptibility of NR, both in the raw or in the vulcanized state, was sufficiently examined and reviewed [5,7,12,21,22]. In another study, rubber-degrading bacterial isolate was enriched from soil sample and according to biochemical and molecular characterization analysis; it was identified as Achromobacter sp. NRB. The obtained strain was capable of degrading and mineralizing raw natural rubber granules as well as some forms of rubber tire and was able to use it as their sole source for carbon. But in contrast to the growth in common media where carbon source is dissolved, the growth of microorganisms on natural rubber products is very slow and the effect of culture conditions on microbial growth on an insoluble solid substrate like rubber has not been well characterized [23]. In the present study, rubber-degrading bacterial consortium was enriched by different carbon and nitrogen sources like beef extract, yeast extract, ammonium sulphate, ammonium nitrate (nitrogen source), glucose, lactose and sucrose (carbon source). From the experiments it was found that beef extract served as a better nitrogen source that enhances the mineralization of latex at its maximum by the bacterial consortium. Likewise, sucrose was found to be the best carbon source that could enhance the mineralization percentage of natural rubber.
Furthermore, Na-succinate positively affected the rate of rubber degradation. Tsuchii et al. [24] reported a significant increase of rubber degradation in presence of other carbon and nitrogen sources. The presence of NH4Cl and K2HPO4 were essential in degradation of rubber substrate where the first was used as N-source and the second serves as a buffer and phosphate source. In the present study, mineral salts medium used in for mineralization of natural rubber contained yeast extract as N-source which increase the bacterial growth and K2HPO4 used as a phosphate source helped the bacterial consortium to utilize natural rubber within 5 days.
Berekkaa et al. [23] also found out that staining with Schiff’s reagent indicated that the colonization efficiency was influenced with the size of rubber substrate. The colonization efficiency was increased when rubber substrate was applied in small pieces (diameter 1 cm2) while; lower colonization efficiency was recorded in case of large size substrate (diameter 1 cm2). A similar finding was done by Tsuchii and coworkers [24] during the growth of Norcardia sp. 835A on latex gloves pieces. Thus, it was clear that the degradation of rubber was positively affected by the amount of NR granules. It was well known that increase in amount of rubber granules leads to increase in surface area of rubber substrate and hence to higher mineralization rate. While in the present study the bacterial consortium was allowed to react with liquid latex at different concentration and it was found that at the lowest concentration of latex the bacterial consortium was able to give the maximum mineralization. Purpose of the present study was to show the growth of bacterial consortium that varies from 0.10% to 0.50% of natural rubber. By performing gene manipulation and by increasing the days of incubation natural rubber present in the environment could be further mineralized by these economic friendly technologies.
The process of degradation of natural rubber compounds by means of microorganism is an eco-friendly manner of controlling pollution in the environment. These microbes can be obtained from anywhere like soil as well as water. Many different types of microorganisms are involved in the biodegradation of NR which promotes different methods of degradation. But it is found that it’s difficult to degrade the NR products at usual environment thus further investigations are required in order to enhance the degradation rate even in usual environmental conditions. In this study it was found that soil bacteria are capable of natural rubber degradation. Even an individual bacterium can degrade natural rubber and the bacterial growth can be enhanced by providing other carbon and nitrogen sources. Thus, it is believable and gives hope that artificial rubber materials can also be degraded by the soil bacteria itself and the degradation rate can be elevated with a little incorporation of such carbon and nitrogen sources.
I express my sincere gratitude to Department of Biotechnology, Stella Maris College for providing all the facilities and helping me in successful completion of my project work. It has been a wonderful experience doing this research work under the guidance of Dr. K Veena Gayathri, Assistant Professor in Department of Biotechnology at Stella Maris College. I also express my heartfelt thanks to my parents for their immense blessings, constant moral support, encouragement and unbreakable trust in me. My sincere thanks to every person who had supported me and held out their helping hands directly and indirectly in completing this work.