Research Article - (2016) Volume 6, Issue 6
Abdul-Hussein ZR* and Atia SS
Department of Biology, College of Science, University of Basra, Iraq
Corresponding Author:
Abdul-Hussein ZR
Department of Biology
College of Science, University of Basra
Iraq
Tel: +96440640 015
E-mail: zr692002@gmail.com
Received date: October 21, 2016; Accepted date: December 06, 2016; Published date: Dec 10, 2016
Citation: Abdul-Hussein ZR, Atia SS. Antimicrobial Effect of Pyocyanin Extracted from Pseudomonas aeroginosa. Eur J Exp Bio. 2016, 6:6.
Copyright: © 2016 Abdul-Hussein ZR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Pyocyanin was extracted from environmental Pseudomonas aeruginosa isolated from soil at Basra city, Bacteria was identified by using Vitek-2 system analysis. Pigment was purified after extraction and identified by using thin layer chromatography, spectrophotometric, and GC mass analysis. Antibacterial and antifungal activities of different concentrations of pigment were studied against urinary tract and wound infection bacteria and pathogenic fungi. Results showed that pigment gave a good effect against all bacteria and fungi.
Keywords
Pyocyanin; Pseudomonas aeruginosa; Antimicrobial
Introduction
Pseudomonas aeroginosa is gram-negative rod shaped, asporogenous, and monoflagellated bacterium. It is about 1-5 µm long and 0.5-1.0 µm wide. P. aeroginosa is obligate bacteria that respire aerobically as its optimal metabolism. However it can also respire anaerobically on nitrate or other alternative electron acceptors. Therefore this makes P. aeroginosa very invasive microorganisms and has been found in environments such as soil, water, humans, animals, plants, sewage, and hospitals [1]. One of the applications of P. aeroginosa in biotechnology is its ability to degrade aromatic hydrocarbon such as methybenzenes, which are the by-products of petroleum industries and used as solvents for enamels and paints as well as in production of drugs and chemicals. Methylbenzenes are considered as environmental contaminants that ubieties in atmosphere, underground and soil, and in surface water [2]. P. aeroginosa can also degrade toluene, the simplest form of methylbenzene by the oxidation of methyl group to aldehyde, alcohol, acid, and convert it to catechol. Therefore it can be used in pollution control [3]. Phenazines are redox-action pigments produced by these bacteria. These pigments are involved in virulence and iron acquisition [4]. Pyocyanin is a water-soluble blue green, phenazine pigment produced by active cultures of pseudomonas aeroginosa. Pyocyanin also has antibiotic activity toward different microorganisms [5,6].
Materials and Methods
Soil samples were collected from different region of Basra city and were serially diluted in sterile distilled water. Processed samples were cultured on nutrient agar. The blue green colonies grown on nutrient agar were sub cultured onto nutrient agar and Gram stained. Cultured plate were incubated for 24-48 h at 25-37°C. Bacteria was identified biochemically with catalase, oxidase and then subjected to Vitek 2 system analysis.
Nutrient broth, mineral salt medium, and peptone water medium was used for the production of pyocyanin pigment. Pseudomonas aeruginosa was inoculated into one of the medium following up and grown for 72 h at 37°C. Bacteria were then removed by centrifugation (10,000xg 30 min) and filtration of the supernatant through 0.45 µm filters [7]. After that, 5 ml Pseudomonas culture was taken in a sterilized tube and same volume of chloroform was added. The solution was mixed well by using shaker for 2 min, and again centrifugation at 10,000xg for 15 min. Two distinct layers separated out, in which one was the pigment, the other remaining material of culture. The pyocyanin was removed and stored in screw caped tube at 4°C. Pigment was passed through TLC plates using chloroformmethanol in the ratio (9:1 v/v) as an eluent [8]. For spectrophotometric analysis, pigment was extracted with chloroform followed by centrifugation at 4000-70,000 rpm for 10-15 minutes and the cell debris was removed by a second centrifugation step. Finally the supernatant was transferred to a cuvette for the measurement of absorbance at different scanning range.
Gas chromatography (GC-Mass) analysis of pyocyanin were performed on a shimadzu Qp2010 quadrupole gas chromatography Mass spectrophotometer (GC-Ms) instrument equipped with a carbowax (30 mm × 0.25 mm ID; 0.25 µm film thickness) capillary column (intercut DB5Ms. Japan). 1 µl of the sample was injected into the capillary column. Helium was used as the carrier gas. Injector and detector temperatures were set at 280°C. Injection was performed in split mode (1:30), the column temperature was programed initially at 40°C for 1 min and then increase at a rate of 5° per min at final temperature of 280°C. Pigments were separated at constant pressure (96.1 Kpa) 6 split ratio 30.0, column flow 1.71 ml/min and peaks were identified by comparing the mass spectra with mass spectral database.
Antimicrobial activity
Effect of pigment against pathogenic bacteria from wound (Pseudomonas aeruginosa, Staphylococcus, E. coli) and urinary tract infection (Pseudomonas aeruginosa, Staphylococcus, E. coli and Bacillus) and pathogenic fungi Cryptococcus neoformans, Candida albicans fungi Aspergillus niger and Aspergillus fumigatus were performed by using agar diffusion methods. Data were summarized as mean ± SD. All data were subjected to statistical analysis using one way analysis of variance (ANOVA) by using mini tab program. The differences were considered significant if P< 0.05.
Results and Discussion
Blue green pigment producing colonies were picked up from cultures grown on nutrient agar and subcultured on nutrient agar and gave name (ps), purified colonies were Gram stained, and catalase and oxidase tests examined, in addition to some biochemical tests (Figure 1).
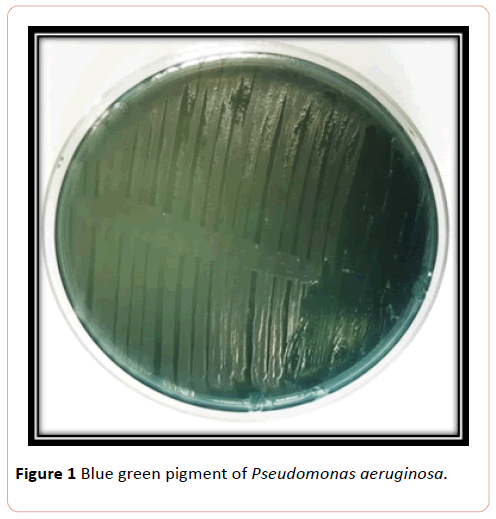
Figure 1:Blue green pigment of Pseudomonas aeruginosa.
Pseudomonas suspected was further diagnosed by subjecting to Vitek system analysis. Results of vitek2 system analysis revealed that bacterial isolates were identified as ps=Pseudomonas aeruginosa (Figure 2).
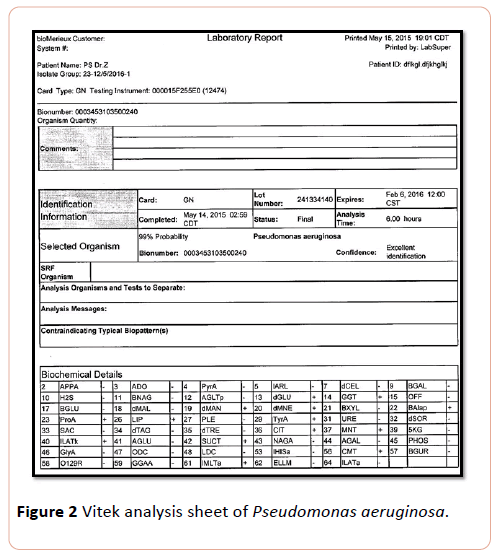
Figure 2: Vitek analysis sheet of Pseudomonas aeruginosa.
Enhancement of pyocyanin production
Different media were used for enhancement of pigment production for pseudomonas (green). Mineral salt medium and peptone water were much better support medium for producing of blue-green pigment, while mineral mannitol, including broth and broth malt with cooked meat extract showed less improvement at same temperature and period of incubation.
The other bacterial pigment was cultured on nutrient agar (NA) and peptone glycerol casein (PGC) gave a good recorded result.
Vitek analysis Pseudomonas aeroginosa
Results of current study showed that isolated pigmented bacteria with greenish color ps exhibited phenomenal probability 99% and confidence excellent identification PS identified as Pseudomonas aeroginosa, this results harmonious with previous studies of who identified different species of bacteria including A. Xylosoxidans, A. baumani, Pseudomonas aeroginosa and P. fluorescens by using vitek. Marko and his colleagues revealed that it can identify different species bacteria isolated from Cf patients and accredit on characteristic features of non-fermenting, Gram negative, Bacilli [9,10].
Extraction and chemical analysis of pigment
Chloroform extracted pyocyanin showed bluish extract converted to red when acidified with 0.1 (N) HCl (Figure 3).
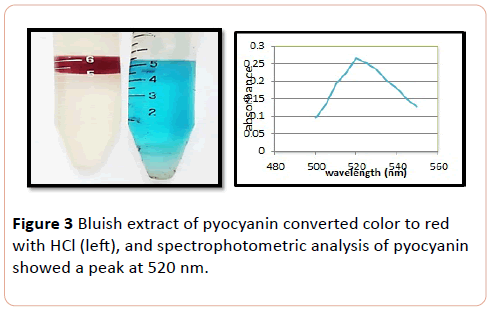
Figure 3: Bluish extract of pyocyanin converted color to red with HCl (left), and spectrophotometric analysis of pyocyanin showed a peak at 520 nm.
GC-MS Chromatograph of Pseudomonas aeruginosa chloroform extracted shown a sharp peak at acquisition time 23.17 minutes on gas Chromatographic analysis which identified by mass spectrum analysis as alpha-Hydroxy phenazine (Hemipyocanin) that gave intense molecular ion peak at 196 m/z with its structure shown in Figure 4.
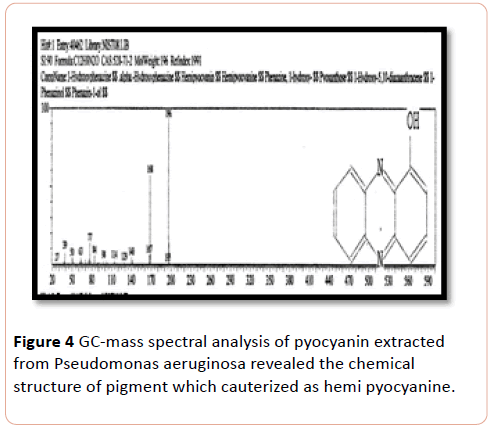
Figure 4: GC-mass spectral analysis of pyocyanin extracted from Pseudomonas aeruginosa revealed the chemical structure of pigment which cauterized as hemi pyocyanine.
TLC
The chromatogram of colored spot on TLC plate showed an RF value of 0.83 for pyocyanin. The result was comparable with previous studies [8]. Baron and John obtained RF value of pyocyanin (0.71) by eluting with chloroform solvent [5]. In the present study pyocanin was separated from culture supernatants by the addition of chloroform solvent. Extractable chloroform layer was mixed with 1 ml of 0.2 (N) HCl which converted pyocanin to pinkish red color which indicated the presence of pyocyanin pigment. The result was consonant with the previous studies [11,12]. Partially purified of pyocyanin was spectrophotometerically scanned at range of 200-800 nm. Absorption was measured at 520 nm, as was also observed by other authors [12,13].
Gas chromatography-Mass of pyocyanin in the present study, showed the presence of Hemipyocyanin and phenazine compound. Previous GC-Ms analysis confirmed the result of the study of who showed the related hemi-pyocyanin pigment extracted from Pseudomonas aerogenosa and identified by its electron impact mass spectrum after gas chromatography at ions at m/z 196 [14].
Antimicrobial effect of pyocyanin
The most affected bacteria to pyocyanin were E. coli extracted from urinary tract, followed by Bacillus and Staphylococcus at the same level. The effect of pyocyanin on wound bacteria was most obvious on Pseudomonas and E. coli, followed by Staphylococcus respectively in the concentration range of 10.000 to 100 µg/ml (P< 0.008) (Figure 5).
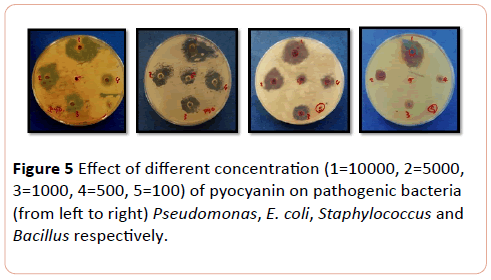
Figure 5: Effect of different concentration (1=10000, 2=5000, 3=1000, 4=500, 5=100) of pyocyanin on pathogenic bacteria (from left to right) Pseudomonas, E. coli, Staphylococcus and Bacillus respectively.
The most affected mould to pyocyanin pigment Aspergillus niger and Aspergillus fumigatus respectively, followed by yeast Cryptococcus neoformans, while Candida tropicalis and C. albicans were equally effected. C. krusi was less affected yeast to (pyocyanin) pigment in the concentration range of 10.000 to 100 µg/ml (P>0.000) (Figure 6).
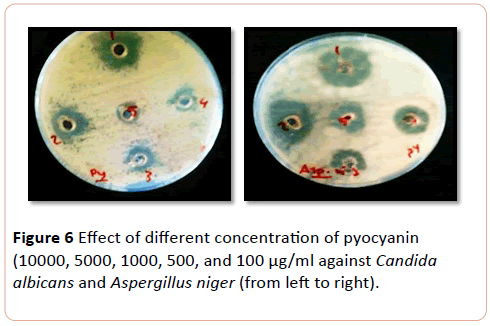
Figure 6: Effect of different concentration of pyocyanin (10000, 5000, 1000, 500, and 100 µg/ml against Candida albicans and Aspergillus niger (from left to right).
Activity of phenazin antibiotics are comparable with other study who revealed phenazin antibiotic have a marvellous effects on Gm+ve and Gm-ve, these results are in agreement with that previous result who reported that phenazin antibiotic have antimicrobial activity only against strains of Bacillus subtilis, Escherichia Coli and Candida albicans [15]. Pyocyanin has been detected in an oil-degrading culture containing Pseudomonas aerogenosa and is a redox-active compound capable of inhibiting the growth of pyocyanin-sensitive of the microbial community [16]. The antagonistic effects of all of phenazine derivatives are ascribed to on general characteristic redoxactivity [17].
Conclusion
Present study concluded that pyocyanin extracted from environmental isolate of Pseudomonas aeruginosa was hemipyocyanin and has antimicrobial effect against infectious bacteria and some pathogenic fungi.