Research Paper - (2017) Volume 7, Issue 5
1Department of Physics, Universidade Estadual Paulista Julio de Mesquita Filho (UNESP), São José do Rio Preto, SP, Brazil
2Department of Biochemistry, Institute of Science, Banaras Hindu University, India
Received date: August 15, 2017; Accepted date: October 06, 2017; Published date: October 16, 2017
Citation: Kumar R, Tiwari S, de Moraes FR (2017) Structure Insights and Fibrinogen Peptides Molecular Recognition by Bothrombin, A Snake Venom Serine Protease from Bothropus jararaca. Eur Exp Biol. Vol. 7 No. 5:32. doi: 10.21767/2248-9215.100032
Copyright: © 2017 Kumar R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Bothrombin is a snake venom serine protease from Bothropus jararaca, which aggregates platelets in the presence of fibrinogen. In this context, the 3D model of Bothrombin was design for molecular modeling studies by modeller9v5 using as template the protein C Activator (PDB ID 2AIP). Energy minimized Bothrombin model was validated by methods such as PROCHECK, PROSA, ERRAT, WHAT-IF and VERITY 3D. After confirming all suggested evidences, such as geometric quality of the backbone conformation, residue interaction, energy profile etc., the interaction study based on docking was further calculated with ClusPro server. This protocol was designed in order to assess the dependence of bothrombin model from its template and how it compares to thrombin. Crystal structure of fibrinopeptides of PDB ID: 1DM4 and 1YCP were selected for docking studies. These fibrinopeptide also docked with thrombin and protein C Activator. All docked complexes were then further analysed by means of interaction energy between subunits and their corresponding binding energy estimates. The present study gain insight into protein structure-function relationships among serine proteinases, and its importance in biomedical applications.
Bothrombin; Inflammation; Biochemistry
Snake venom serine proteases (SVSPs) are most important and commonly used in the present era in the field of drug designing. This protease catalyze the cleavage of covalent bond of peptides and act as important role in diverse biological processes such as digestion, regulation of blood coagulation, inflammation, immunity etc. [1]. This protease are grouped into six major clans and subsequently divided into families based on their sequences (MER-OPS classification, https:// merops.sanger.ac.uk; [2]: exclusively clan SA, family S1. Among all of them, SVSPs display specifically biological reactions in the blood coagulation cascade and platelet activation process. Physiologically, SVSPs activate the prothrombin and subsequently shorten the coagulation times. However, some SVSPs also possess fibrinogen-clotting activity and act as thrombin-like enzymes such as bothrombin, SVSP from Bothropus jararaca.
Bothrombin was crystallized by the same lab [3], though no three dimensional structure was made available so far. Moreover, theoretical model of Bothrombin (1CXM), a serine proteinase with fibrinogenolytic activity from the venom of Bothrops jararaca was published by Serrano et al. [4]. Bothrombin form aggregates platelets in the presence of fibrinogen and interact with glycoprotein 1b, which activates blood cogulation factor VIII. Physiologically, it selective cleave Arg-|-Xaa bond in fibrinogen, to form fibrin and release fibrinopeptide A [5]. Thus, this enzyme encourages us to do docking analysis with fibrinopeptide and get information about its precise role in the blood coagulation process. This enzyme is highly endemic and that is the reason many industries and academic groups are looking for its inhibitor since the last decade. The treatment usually falls into inhibitor such as diisopropyl fluorophosphates but limited therapies due to serious and life threatening side effects. At present, modern branches such as biochemistry, molecular biology, genetics and pharmacology have grown significantly in the ability to identify specific biological targets [6]. In this continuation, computational tools may add some fast and low-cost input to make data more important to explore such targets in designing new drugs with the aim to decrease fatality. Molecular docking and molecular dynamics are two widespread used computational tools for gaining insight into protein targets and their interactions and dynamics at molecular level.
However, it has already been mention that bothrombin act as thrombin like activity but drug interaction studies about bothrombin inhibition assay are not found in the literature [7]. In such case, fibrinopeptide with PDB code; 1DMK and 1YCP has been used for docking study with bothrombin, which are 11 and 23 amino acid respectively [8].
In the present study, the 3-D structure of bothrombin has been constructed using resulting FASTA sequences. Further, the molecular model of bothrombin was constructed using Modeller9v5 package for homology modeling using template 2AIP; protein C activator. The resulting model quality was checked by using PROCHECK, PROSA, ERRAT, VERIFY 3D and WHAT-IF and as a result on the overall scores, we choose the final model. After final confirmation, protein-protein docking was performed between the molecular models of bothromin and fibrinopeptide (1DMK and 1YCP) by cluspro software. The protein-protein docking analysis has also been done with these peptides with protein C activator (2AIP) and thrombin (1BTH) crystal molecule. At last, the binding energy in between all complexes has been calculated by tinker software and analysis the interpretation. The complete analysis of apparent inhibition in addition to interaction of the models was carried out with high binding affinity; make us inform that bothrombin act as thrombin with more or less same catalytic activity. The resulting result presented in this article will be useful to design molecules for drug designing.
All the estimations were execute on a workstation Hi-end server: Intel core 2 duo processor, 32 bits with 2 GB RAM with video graphics card. Molecular modeling tasks were achieved with Modeller9v5 (REF); protein-protein docking computations were carried out with ClusPro server (REF). For homology modeling of the Bothrombin, the crystal structure of Protein C activator (PDB ID: 2AIP) from the venom of copperhead snake Agkistrodon contortrix was used as a template and the docking had been done with two fibrinopeptide; (1) PDB ID: 1DM4 chain C (2) PDB ID: 1YCP chain F, with all default settings during calculations.
Sequence alignments
Sequence of Bothrombin (gi No. AAB30013.1) was obtained from National Centre for Biotechnology Information (NCBI). The target sequence of bothrombin is used as a query sequence to search for homologous protein structure(s) that could serve as template(s) by running pblast [9] against PDB [10]. By using blastp program template sequences of low similarity were obtained. In order to enhance the query coverage we opted for combination of multiple templates. Although the idea of combining multiple templates sounds straightforward, its implementation is fairly complex. The real challenge is not the identification of a list of suitable template candidates, but an optimal combination of these. This is because template search methods ‘outperform’ the needs of comparative modeling in the sense that they are able to locate so remotely related sequences for which no reliable comparative model can be built. The reason for this is that sequence relationships are often established on short conserved segments, while a successful comparative modeling exercise requires an overall correct alignment for the entire modeled part of the protein. To find out an appropriate template structure Protein C activator; 2AIP (gi No. AAO85513.1) was selected, which has maximum identity. The BLASTp alignment between selected templates was further refined using sequence alignments in the ClustalW2 (EMBL-EBI) online software with default parameters [11].
Molecular model building
The search using the BLASTp alignment algorithm within the PDB database showed various potential templates for molecular modeling purposes. Only limited, crystallographic structures were found to show high identity score with respect to bothrombin sequence. Among all of them, PDB ID (2AIP) shows maximum identity, this would be the reason; 2AIP was selected as template for bothrombin for further studies. This sequence was further refined using sequence alignment in the ClustalW 2.0.12 with default parameter. However, the 3D structure of bothrombin was predicted by homology modeling on the basis of the structure of by using the program Modeller 9v5. Modeller 9v5 is used to develop the structures for the proteins with unknown structure by using comparative homology [12]: https:// salilab.org/modeller/tutorial/advanced.html]. A number of models were generated by Modeller9v5 and were visualized using Pymol. The modeling procedure begins with an alignment of the sequence to be modeled (target) with relative known three-dimensional structure (template). Many models were generated and among them the one having lowest root mean square deviation (RMSD) value when superimposed onto the template 2AIP was chosen for further studies.
Validation of the homology model
After the construction of the model, its quality was assessed considering both geometric and energetic aspects using PROCHECK [13], ERRAT [14] and PROSA [15] for internal consistency and reliability. The Ramachandran plot computed with PROCHECK provided the residue position in particular segment based on the dihedral angles. Finally, the best-quality models were subjected to further calculations and molecular modeling studies, binding site analysis and other calculations.
The structurally conserved regions (SCRs) between reference proteins were then identified and superimposed, and then the chosen models were subjected to energy minimization in order to obtain a stable, low-energy conformation. We had generated five models by Modeller, were further evaluated for quality by calculating their energies. On evaluating the energies of these models, Model 2 was found to have least energy value (DOPE score) -14313.526367 (data not mention). This predicts the good quality of Model 2 with relatively compact structure.
Protein-fibrinopeptide docking and analysis
In order to get insight into possible protein-fibrinopeptides interactions, we have used six complexes formed between thrombin (PDB code 1BTH), Protein C activator (PDB code 2AIP) and energy minimized bothrombin with two fibrinopeptide (PDB codes 1YCP and 1DM4). The docking was performed by homology to the known complex conformation available in the PDB by using PyMOL structural alignment tools. Following, the initial complex coordinates were subject to energy minimization in GROMACS.
To further compare each protein-peptide complex, the electrostatic potential complementarity between the proteininhibitor complexes were also checked using the pdb2pqr web server [16] and APBS software [17]. The electrostatic potential surface were generated and compared to the experimentally solved thrombin and protein C activator.
Binding affinity
To further compare the studied complexes, we assessed the energetics of each resulting protein-peptide complex. The TINKER Molecular Modeling Package [18] was used to energy minimize each structure (monomers subunits and complexes) and to compute the binding energy components. By using the same model as REF, we state that the binding energy may be estimated by:
ΔGbind=Gcomplex-Gprotein-Gpeptide
Where the thermodynamic potential G may be approximate to the bonding, angle, van der Waals and Coulombic terms:
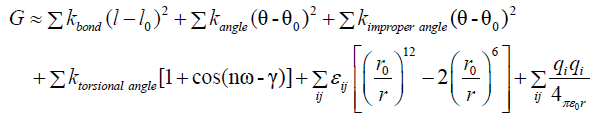
Where each constant are taken for the selected force field in TINKER Molecular Modeling Package. For this study we used the AMBER force field [19].
Virtual alanine scanning
In order to get a more detailed picture of the most important residues for establishing the protein-protein interaction interface, we used computational alanine scanning. Robetta Alanine Scanning [20,21] was employed for such a task. In this protocol, each amino acid residue is replaced by alanine residues. The difference in the binding energy (as calculated by Robetta Alanine Scanning algorithm) is then associated to the amino acid importance. Amino acid residues which contribute with more than 2 kcal/mol for the protein-protein complex binding energy is referred to as hot spot.
Sequence alignments
To attain an appropriate template structure for assembling the target pattern, the sequence of bothrombin was retrieved from NCBI (National Center for Biotechnology Information). To get a high-quality template for homology modeling of bothrombin, the amino acid sequence was aligned against the Protein Data Bank [22]. This alignment was executed by means of the BLAST algorithm. Subsequently, further refinements of these sequences alignment in the ClustalW 2.0.12 with default restrictions were performed. Multiple sequence alignment informs that the 2AIP and bothrombin are 71% similar in nature and have major difference in the amino acid sequence on the turn and bend region as shown in Figure 1, performed by ClustalW 2.0.12. This information also support that 2AIP was selected as good template for modeling of Bothrombin. The figures informs that the residues involved in binidng of various feedback inhibitors in template were also been conserverd in the Bothrombin (data not shown).
Molecular model building
The exploration by the BLASTp alignment algorithm within the PDB database illustrates impending templates for molecular modeling objects. Selection of appropriate template was chosen based sequence similarity, residue compactness, and crystal resolution [23]. Many structures were found to show high identity score with respect to bothrombin but among them, crystal structure of native protein C activator (2AIP) from the venom of copperhead snake Agkistrodon contortrix (PDB code: 2AIP, resolution 1.65 Å, R-value 0.17086 (obs), and R-Free 0.19692) [16] was selected as template structure. The 3D structures bothrombin was predicted by homology modeling on the basis of the structures of 2AIP by using the program Modeller 9v5. This software is an automated attempt to comparative modeling, begins with an alignment of the sequence to be modeled as target with relative known threedimensional structures act as templates. Many models were generated and among them the one having lowest root mean square deviation (RMSD) value was selected as template for further analysis [24]. At last, the chosen models were subjected to energy minimization to get the most stable conformation for our study.
Validation of the homology model
The quality of the constructed model was evaluated by both geometric and energetic aspects using PROCHECK, ERRAT and PROSA for internal uniformity and consistency. The first validation was carried out using Ramachandran plot analysis computed with PROCHECK. PROCHECK has been used to provide residue position based on the dihedral angles in the form of Ramachandran plot by checking residue-by-residue stereochemical quality of the protein structure [25]. The analysis showed that 2AIP, energy minimized Bothrombin and theoretical model of Bothrombin (1CXM) in the most favorable region were 83.3%, 78.2%, and 74.1% and in the additional allowed region 15.6%, 17.6%, and 22.8%, respectively (Table 1 and Figure 2A and 2B).
| Structure | Average package qualitya | Rotamer normalityb | Backbone conformation | Bond lengthc | Angled |
|---|---|---|---|---|---|
| 2AIP | 0.086 | 0.539 | -5.278 | 0.478 | 0.718 |
| Bothrombin | -0.54 | -2.555 | -6.319 | 0.67 | 1.233 |
| 1CXM | -0.82 | -1.975 | -10.199 | 1.234 | 1.419 |
| 1BTH_H | -1.418 | -1.637 | -5.636 | 0.475 | 0.753 |
(a) The average quality of 200 highly refined X-ray structures was -0.5 ± 0.4
(b) The behaviour of the distribution is much that a Z-score below -2 (2 standard deviations way from the average) is poor and a Z-score of less than -3 is of concern; positive is better than average
(c)RMSD Z-score should be close to 1.0
(d)RMSD Z-score, more common values are around 1.55
Table 1: What if stereochemical quality evaluation.
Figures 2A and B: Ramachandran plot of developed. Ramachandran plot by checking residue-by-residue stereochemical quality of the protein structure. The analysis showed that 2AIP, energy minimized Bothrombin and most favorable region were 83.3%, 78.2%, and 74.1% and in the additional allowed region 15.6%, 17.6%, and 22.8%, respectively.
PROCHECK has been used to provide the best-quality models, which were selected for further molecular modeling studies, binding site analysis and some other necessary information. The structurally conserved regions (SCRs) between suggested proteins were then recognized and superimposed as a result the bothrombin sequence was then aligned to the SCRs using the alignment module. The superimposition of 2AIP and bothrombin has shown here in Figures 3A and 3B where final modeled structure of bothrombin is composed of 13 beta-strands arranged into two domains, similarly to 2AIP structure, and two long alpha-helixes and short one near the catalytic site (Figure 4). The catalytic region lies on the surface between the two beta-strands motifs. The catalytic triad (His 57, Asp102, and Ser195) formed in the 2AIP have similar orientation as compare to the catalytic residues His-41, Asp-86 and Ser-178 of bothrombin are highlighted in Figures 5A and 5B. The superimposition of bothrombin did not show major structural conformational changes in comparison to the template model; 2AIP, which in turn is consistent with the relatively low RMSDF values. The overall low RMSD values reflect the high structural conservation of this complex through evolution, making it a good system for homology modeling (Table 2).
| PROCHECK | Z score | ERRAT Score | |||||
|---|---|---|---|---|---|---|---|
| Ramachandran plot quality (%) | Goodness factor | ||||||
| Most favored | Additional allowed | Generously allowed | Dis-allowed | Overall | |||
| 2AIP | 83.3 | 15.6 | 1 | 0 | 0 | -6.57 | 83.019 |
| Bothrombin | 78.2 | 17.6 | 3.1 | 1 | -0.4 | -6.7 | 80.097 |
| 1CXM | 74.1 | 22.8 | 2.1 | 1 | -0.6 | -7.11 | 55.051 |
Table 2: PROCHECK and ERRAT.
By applying the structural superposition and RMSD evaluations, our model appears very similar to the theoretical model, 1CXM but with good quality assessment analysis. The total quality G-factor -0.4 was calculated (average value are in between 0 and -0.5), which informs that the model was good in structure. However, G-factor value of 1CXM was -0.6; consider being not a good model for further analysis.
However, PROCHECK analysis showed no bad scores for side chain and main chain parameters for both bothrombin and 1CMX, but has better quality for bothormbin. In addition for non-bonded atomic interactions, ERRAT score is consider as overall quality factor and higher value (>50) consider as better quality. In the current case, the ERRAT score for bothrombin was 80.097, which fit well within the high quality model as shown in Figure 5A. However, the ERRAT score of 1CXM was 55.051, which in comparison to support that the bothrombin model is more perfect for further analysis as shown in Table 1.
In the analysis, PROSA Z-score was used to judge the interaction energy of each residue with the remainder of a protein was computed whether it fulfills certain energy criteria or not. Evaluation of the energy minimized model of bothrombin with PROSA web revealed that Z-score value was -6.7 as shown in Table 1 and Figure 5B. This indicates no significant deviation from typical native structure as the template when compared with Z-score of -6.57 for 2AIP template. However, the Z-score of 1CXM was -7.11, informs that modeled bothrombin have better homology with 2AIP in comparison to 1CXM. Thus the bothrombin structure was found to be reasonable and reliable conformation for further protein docking studies.
Protein-protein docking
I need these PDBs of the complexes that will be analyzed. In order to elucidate the inhibition mechanism on bothrombin, two fibrinopeptide (1dm4 and 1ycp) were used as a ligand for docking study to evaluate the productive ability for use in the computational structure-based protein-protein studies. As reported, bothrombin behave like thrombin but due to limitation of its structure, we performed modeling of bothrombin by using fasta sequence as reported. Fasta sequence of bothrombin has similarity (more or less similar RMSD) with 1BTH and 2AIP as mention in the (Figure 6A), encourage us to select thrombin as a template. But, among both of them 2AIP have high resolution (1.65 Å) than 1BTH (2.30 Å), which give it more preference. The electrostatic potential surface as shown in Figure 6B, inform that active site of bothrombin have negative charges surrounded by positive charges as similar in case of 1BTH rather than 2AIP. This information encourage us that botrombin would behave like thrombin and have play important role in degradation of fibrinogen. However, as shown in Figure 6C, the active site pocket of bothrombin is more similar to 2AIP than 1BTH, informs bothrombin have similar selection of substrate as 2AIP. This all information motivates to do docking of bothrombin, 2AIP and 1BTH with fibrinopeptide (1YCP and 1DM4). To perform the protein-protein docking between all complexes, ClusPro server was used in its default parameters, and potential attractions between the interface residues, as assessed in the experimentally solved complex were employed. The ten best representatives of different clusters were compared, achieving on average 1 Å RMSD and up to 5 Å RMSD (Choose a threshold less than 5A (2 or 3A I think) so that it has only 4 structures for each case, it would be very expensive experimentally to test 10 cases for each protein-ligand). Further, all six complexes (2AIP_1DM4, 2AIP_1YCP, 1BTH_1DM4, 1BTH_1YCP, Bothrombin_1DM4 and Bothrombin_1YCP) are further use for binding energy calculations as shown in Figure 7A-7C. This informs that 2AIP_1DM4 and 2AIP_1YCP have less ΔG value in comparison to rest of the 1DM4 and 1YCP complexes, respectively (Table 3).
| Bond Stretching | Angle Bending | Improper Torsion | Torsional Angle | Van der Waals | Charge-Charge | Implicit Solvation | Total Potential energy | Intermolecular Energy | |
|---|---|---|---|---|---|---|---|---|---|
| Bothrombin | 322.2461 | 708.8676 | 34.6025 | 2295.1609 | -838.9149 | -7299.3628 | -1375.505 | -6152.9056 | |
| 2AIP | 166.7472 | 499.3745 | 16.3666 | 2026.5276 | -1246.6104 | -8740.7519 | -2943.3916 | -10221.7381 | -1617.7321 |
| 1BTH | 157.6084 | 516.6546 | 17.5501 | 2181.1625 | -1480.1138 | -6998.2166 | -4901.7227 | -10507.0775 | -454.3891 |
| 1DM4_C | 7.0249 | 11.7386 | 0.3714 | 54.9343 | -15.9343 | -421.0023 | -270.2253 | -633.0875 | -32.0757 |
| 1YCP_F | 5.365 | 11.0937 | 0.1313 | 35.9067 | 1.5188 | -289.9766 | -213.2131 | -449.1744 | -31.4309 |
| 1BTH_1DM4 | 189.0033 | 650.0719 | 42.9134 | 2439.375 | -1247.2866 | -10403.9477 | -2279.6641 | -10609.5347 | -663.0243 |
| 1BTH_1YCP | 1382.9255 | 684.9666 | 47.982 | 2440.9696 | -641.9088 | -9735.8124 | -2738.7649 | -8559.6423 | -625.1846 |
| 2AIP_1DM4 | 190.1015 | 544.5895 | 32.58 | 2206.5655 | -1017.0955 | -11395.7499 | -814.3604 | -10253.3693 | -1665.63 |
| 2AIP_1YCP | 191.1646 | 537.8985 | 32.5679 | 2189.6982 | -980.847 | -10878.0231 | -1143.7864 | -10051.3273 | -1606.4641 |
| both_1DM4 | 131.493 | 587.26 | 42.0334 | 2307.9445 | -1259.988 | -7993.655 | -1279.9834 | -7464.8964 | -135.4541 |
| both_1YCP | 133.4075 | 595.3572 | 38.0078 | 2281.1657 | -1216.1594 | -7790.8477 | -1351.1726 | -7310.2415 | -97.3496 |
Table 3: Binding energy calculation of different complexes of docking studies.
The BLASTp result revel that bothrombin sequence has high sequence and structure similarity with 2AIP, which depicts that, the conserved amino acids intricate in the binding of active site catalytic triad (His 57, Asp102, and Ser195). The catalytic site region as shown as bobble are also shown in the Figure 6, informed that bothrombin share the same pocket as 2AIP. The bothrombin also have similar electrostatic potential with 1BTH (thrombin) as shown in Figure 6, inform that it would behave as thrombin like protein and act as role in degradation of fibrinogen. All these proteins viz, bothrombin, 2AIP, 1BTH were share lowest RMSD value, which finally select bothrombin as model for this study. Bothrombin was modeled by Modeller 9v5 and the final selected bothrombin model, the energy minimization was calculated by Tinker software to elucidate the most stable conformation for study. The quality of the model was appraised by PROSA, PROCHECK, ERRAT, and WHAT IF for their consistency and stability as also reported. The PROSA was used to calculate the interaction energy per residue and evaluate whether it fulfills certain energy criteria or not. The PROSA Z-Score is their output result, which analyzed. ERRAT on the other hand consider as overall quality factor and its higher score mean better quality.
As reported, the bothrombin ERRAT score is more than 80.097, which inform that the consider model is one of the best one. The model has high similarity with theoretical model 1CXM after calculated by PROCHECK analysis as shown in Table 1 and Figure 2A and 2B. All information suggests that the bothrombin behave as thrombin and to do further docking with fibrinopeptide were analyzed as reported. The results were shown in Figure 7, which informed that 2AIP, 1BTH and bothrombin are able to degrade the fibrinopeptide. Similar analysis with three dimensional interactions also reviewed.
To elucidate the interaction as wet lab would be a very expensive process, in such context; this study wills empowerment to biochemist and pharmacologist to use bothrombin as one of the substitute of Thrombin in case of fibrinolytic degradation. The generated bothrombin model is anticipated to be useful for structure based drug designing, in addition.